LowHweeMeng_FYP
advertisement

SIM UNIVERSITY
SCHOOL OF SCIENCE AND TECHNOLOGY
DEVELOPMENT OF A COMPUTATIONAL
MODEL FOR CALPAIN CLEAVAGE SITES
PREDICTION
STUDENT
: LOW HWEE MENG (Z0704443)
SUPERVISOR : WEE JIN KIAT, LAWRENCE
PROJECT CODE: JUL2010/BME/039
A project report submitted to SIM University in partial fulfillment of the
requirements for the degree of Bachelor of Biomedical Engineering
May 2011
Page | 1
Acknowledgement
I would like to express my gratitude and thanks to the following people who have
made this Capstone Project possible:
• Dr Wee Jin Kiat, Lawrence, for his supervision, support and advice over the course
of the Capstone Project.
• My parents, loved ones and friends for their patience and support.
• My team leader, Mr. Thoreau Hervé and fellow colleagues at the Genome Institute
of Singapore, Genome Technology and Biology department for their patience and
understanding.
Page | ii
Table of Contents
Page
Acknowledgement
ii
Table of Contents
iii
List of figures
vi
List of tables
vii
Abstract
viii
Part 1
Chapter 1: Calpain……………………………………………………………………1
1.1. Calpain discovery and biology……........................…………………………1
1.2. Calpain superfamily and structure…………………………………………...2
1.3. Calpain and disease implication..……………………………………………9
1.3.1. Role of calpain in apoptosis
1.3.2. Role of calpain in neural degeneration
1.4. Challenges in deciphering protease cleavage……………………...………...12
1.5. Project objectives…………………………………………………………….13
Chapter 2: Computational approaches to data classification ………………………...15
2.1 Introduction to Support Vector Machines (SVM)…………………………...15
2.2 Current perspective in calpain cleavage prediction………………………….16
2.2.1
Sequential determinants of calpain cleavage
2.2.2
Group-based Prediction System-Calpain Cleavage Detector
(GPS-CCD)
2.2.3
CaMPDB: a resource for calpain modulatory proteolysis
2.3 Summary.........................................................................................................19
Page | iii
Chapter 3: Calpain dataset……………………………………………………………20
3.1 Dataset collection…………………………………………………………....20
3.2 Data extraction and cleaning………………………………………………...22
3.3 Summary..........................................................................................................23
Chapter 4: Prediction of Calpain Substrate Cleavage………………………………...24
4.1 Introduction…………………………………………………………………..24
4.2 Materials and Methods……………………………………………………….24
4.2.1
Calpain datasets
4.2.2
Symmetrical subsequence extraction
4.2.3
Asymmetrical subsequence extraction
4.2.4
Training and test dataset
4.2.5
Vector encoding schemes
4.2.5.1 Simple binary encoding
4.2.5.2 Bayes Feature Extraction (BFE) encoding
4.2.6
SVM implementation
4.2.7
SVM optimization
4.2.8
SVM training and testing
4.2.9
Linear sequence analysis of primary calpain dataset
4.2.9.1 Relative position-specific amino acid propensity
4.2.9.2 Sequence logo representation of calpain cleavage events
4.3 Results and discussion……………………………………………………….37
4.3.1
Performance metrics of SVM prediction
4.3.2
Relative position-specific amino acid propensity
4.3.3
Sequence logo representation of calpain cleavage events
Page | iv
Chapter 5: Prediction of Receptor Tyrosine Kinases (RTKs) Family Proteins……….49
5.1 Introduction to Receptor Tyrosine Kinases (RTKs)………………………….49
5.2 Prediction of calpain cleavage of RTKs……………………………………...51
5.3 Summary...........................................................................................................52
Chapter 6: Conclusion………………………………………………………………....54
6.1 Summary of project report…………………………………………………....54
6.2 Recommendations and future direction……………………………………....56
Part 2
Chapter 7: Critical reviews and reflections……………………………………………58
REFERENCES………………………………………………………………………..61
Appendix A……………………………………………………………………….......65
Appendix B…………………………………………………………………………...75
Appendix C…………………………………………………………………………...80
Appendix D…………………………………………………………………………...83
Appendix E…………………………………………………………………………....87
Page | v
List of figures
Page
Figure 1-1: Schematic structures of calpain superfamily members
across various organisms..............................................................................................4
Figure 1-2: Domain structure of the human calpain family………………………….6
Figure 1-3: Crystallographic structure of human m-calpain…………………………7
Figure 1-4: Schematic representation of calpain activation in various
neurodegenerative diseases…………………………………………………………..11
Figure 2-1: Illustration of SVM concepts……………………………………………15
Figure 3-1: A summary of the calpain dataset construction process………………...23
Figure 4-1: Symmetrical subsequence segments extracted for
SVM training and testing……………………………………………………….........25
Figure 4-2: Asymmetrical subsequence segments extracted for
SVM training and testing………………………………………………………….…26
Figure 4-3: A schematic representation of datasets used for SVM training
and testing…………………………………………………………………………....28
Figure 4-4: Flowchart of SVM workflow…................................................................32
Figure 4-5: Graphical representation of the trends in SVM classifiers
performance in terms of A) accuracy and B) AROC scores for various
subsequence windows…………………………………………………………….….39
Figure 4-6: Heatmaps of position-specific amino acid intensities of
A) positive examples, B) negative examples and C) calculated propensity Px...........44
Figure 4-7: Sequence logo representation of experimentally-verified
calpain cleavage events………………………………………………………………45
Figure 5-1: Construction of the 40-mer moving window in
EGFR (P00533). …………………………………………………………………….51
Page | vi
List of tables
Page
Table 1-1: Members of the calpain family, encoding genes
and associated polypeptides………………………………………………………….8
Table 4-1: Summary of SVM prediction performance of classifiers trained
using various subsequences and encoding strategies……………………...........…...37
Table 4-2: Comparison of position-specific amino acid prevalence
in the generated 40-mer sequence logo versus findings by Tompa et al……………47
Table 5-1: Schematic maps of predicted calpain cleavage sites
on the receptor tyrosine kinase (RTK) family subset………………………………..53
Page | vii
Abstract
Calpains constitute an important family of calcium-dependent cysteine proteases
widely expressed in mammalians and conserved across eukaryotes. Distinguished by
limited proteolysis of protein substrates at neutral pH, calpains modulate key
biological processes such as apoptosis, cytoskeletal organization and neuroendocrine
pathways. Aberrations of calpain function are known to be implicated in cancers and
neurodegeneration. Despite numerous efforts to unravel calpain regulatory roles, the
precise mechanisms of substrate recognition and calpain-dependent cleavage have not
been fully established. Recent development of calpain cleavage sites prediction
methods achieved varying degrees of success and revealed interesting observations to
amino acid sequence conservation and asymmetrical contributions of amino acids to
calpain substrate recognition. A set of 341 unique calpain substrate cleavage sites
were obtained from available databases and literature searches and analyzed. To
determine unique sequence features in calpain substrates, linear sequence analysis via
sequence logo and heatmap generation as well as derivation of amino acid propensity
was conducted and revealed correlation to previous sequential studies and also
significant propensity for alanine, tryptophan, methionine, proline and serine residues
within the P4-P4’ window and downstream regions of cleavage sites. Next, to
investigate the efficacy of developing a support vector machine (SVM)-based method
for calpain cleavage site prediction, a series of SVM classifiers designed to
encapsulate the cleavage sites with various extracted subsequences (symmetrical
P4P4’, P8P8’, P12P12’, P16P16’ and P20P20’ and asymmetrical P4P12’ and P12P4’), together
with a combined approach of simple binary and bi-profile Bayes Feature Extraction
(BFE) encoding were implemented and evaluated. Predictive performance of the
SVM method achieved an accuracy ranging from 71% to 86% with AROC score
Page | viii
ranging 0.788 to 0.927 on independent test sets, with significant improvement in
overall performance with BFE encoding and longer subsequence windows.
Application of our best performing prediction model on a subset of receptor tyrosine
kinases (RTKs) revealed potential calpain regulation and involvement in the apoptosis
cascade as effectors of survival and growth signals. This study has presented an SVMbased approach for calpain substrate cleavage site prediction, highlighting its potential
to complement experimental efforts to elucidate calpain cleavage mechanisms and
degradome. The content of this project has been accepted for poster presentation in
the 19th Annual International Conference on Intelligent Systems for Molecular
Biology and 10th European Conference on Computational Biology (ISMB/ECCB),
Vienna, 2011.
.
Page | ix
PART 1
Chapter 1: Calpain
1.1
Calpain discovery and biology
Proteases play an important role in the regulation of biological functions in the body.
Calpains (EC 3.4.22.52/53) constitute an important family of intracellular, calcium
(Ca2+) dependent, non-lysosomal cysteine proteases which exhibit limited proteolytic
activities at neutral pH, in contrast to complete digestion. Calpains and its numerous
homologues form a major protease family widely expressed in mammalians and
organisms such as plants, bacteria, yeast and fungi, seemingly conserved across
eukaryotes. Limited proteolysis by calpains alters substrate structure, leading to
regulation of biochemical activities and cellular functions, deeming calpains as
“intracellular modulators”.
Calpains is involved in important biological processes such as programmed cell death
(apoptosis), cytoskeletal organization and neuroendocrine secretory pathways.
Numerous calpain substrates are localized to the cytoskeleton and secretory pathway
proteins, which affects cell structure, shape and cellular interactions. Cytoskeletal
degradation may cause disruption to secretory pathway dynamics causing
accumulation of large intracellular protein aggregates from proteolytic end-products.
Calpain involvement in cytoskeletal protein proteolysis has been associated to
neuronal diseases and their pathology in Huntington’s, Alzheimer’s and Parkinson’s
disease (Evans, et al., 2007).
Page | 1
Initial discoveries of calpain were reported in the 1960s, from calcium-dependent
proteolytic events detected in rat brain (Guroff, et al., 1964) and skeletal muscles
(Ishiura, et al., 1978). These events were attributed to “calcium-activated neutral
proteases” or CANP due to calcium requirements and activity at neutral pH. In the
same study, Ishiura, et al. also achieved the purification of the CANP molecule into
homogeneity.
The first study on cDNA cloning of the calpain catalytic subunit gave structural
evidence of a chimeric molecule consisting of a cysteine protease, similar to papain
originating from papaya, and a calmodulin-like Ca2+ molecule, with calmodulin being
a calcium-regulated signaling protein, leading to its initial nomenclature of “calpain”
(Ohno, et al., 1984). A nomenclature review of calcium-dependent proteinases unified
CANP and “calpain” to calpain (Suzuki, 1991). Calpain is classified under the papain
superfamily, which includes Clan CA, family C1 and C2, forming three distinct
families, namely bleomycin-hydrolase (BLH)-type, papain–type and calpain-type
(Berti and Storer, 1995).
1.2
Calpain superfamily and structure
The calpain system comprises of three molecules, two calcium (Ca2+) dependent
proteases, -calpain (calpain 1) and m-calpain (calpain 2) and calpastatin, a highlyspecific inhibitor of both - and m-calpains. Being the best characterized members of
the superfamily, - and m-calpains are referred to as “classical” calpains, with and
m referring to the micromolar and millimolar Ca2+ requirements in-vitro for protease
activity respectively. Both - and m-calpain consists of two distinct subunits, an 80-
Page | 2
kDa large catalytic subunit and a 28-kDa regulatory subunit, together forming a
heterodimer. The large subunits (μCL in -calpain and mCL in m-calpain) are nonidentical, however, sharing a 55-65% amino acid sequence homology (Goll, et al.,
2003).
Numerous genomic studies in the past two decades have led to the discovery of
hundreds of calpain-related homologues in various organisms, contributing to a
superfamily of versatile functions. In humans, fifteen genes have been discovered to
encode calpain-like protease domains, generating a diverse range of homologues with
varying functional domain combinations. Figure 1-1 depicts the schematic
representation of calpain superfamily members and homologues.
Page | 3
Figure 1-1: Schematic structures of calpain superfamily members across various organisms.
Page | 4
(Adapted from CaMPDB- Calpain for Modulatory Proteolysis Database)
Deciphering calpain superfamily domain structure is essential in understanding calpain
structure-function relationship. Calpains can be classified in two general groups; typical
and atypical calpains (Figure 1-2). Typical calpains (1, 2, 8, 9, 11, 12 and 14) consists of
four well-established domain structures: domain I (autolytic activation); domain II
(cysteine catalytic site, constituting active sites IIa and IIb); domain III (C2-like Ca2+binding sites) and domain IV (calmodulin-like Ca2+ -binding sites, resembling the pentaEF hand family of polypeptide). An exception is calpain 3 (skeletal muscle-specific
calpain, p94), possessing three additional characterizing regions, NS, IS1 and IS2 (Strobl,
et al., 2000; Hosfield, et al., 2001).
Atypical calpains (5, 6, 7, 10, 13, and 15) are monomeric calpains, lacking the
calmodulin-like penta-EF hand sequences in domain IV. Instead, calpain 5, 6 and 10,
possess a C.elegans, TRA-3 like T-domain (Dear, et al., 1997; Horikawa, et al., 2000).
Calpain 7 possesses a large N-terminal domain, together with a PalB homologous Cterminal domain resembling the PalB protease originating from the A.nidulan (Franz, et
al., 1999). Calpain 15 was observed to be a vertebrate homolog of the D.melanogaster
small optic lobe gene (SOL), with high homology at the catalytic and C-terminal domains
(Kamei, et al., 1998).
Page | 5
Figure 1-2: Domain structure of the human calpain family.
(Adapted from Evans, et al., 2007)
As discussed earlier, classical calpains possess large catalytic subunits, μCL and mCL,
encoded by the CAPN1 and CAPN2 genes respectively. The calpain small subunit
encoded by the CAPN4 gene (calpain 4) consists of two domains, V and VI, and common
to both calpain 1 and 2, (Franco and Huttenlocher, 2005). Figure 1-3 shows the
crystallographic structure of human m-calpain made up of large subunit mCL domains
and small regulatory subunit domains. Other typical calpains share the similar large
subunit domain structure as classical calpains; however, they do not form a heterodimers
with the small subunit.
Page | 6
Domains dI, dIIa, dIIb, dIII, dIV, dV
and dVI are labeled in different colors.
I-II is the α-helix linking domains I and
IIa. The linker domain is represented
by a red line running from the gap
between dIII and dIV to the bottom
right of the diagram labeled III-IV.
The active sites, cysteine, Cys-105;
Histidine, His-262, Asparagine, Asn286 and Tryptophan, Trp-288 are
highlighted in gray at the top of the
domain IIb.
(Adapted from Reverter, et al., 2001)
Figure 1-3: Crystallographic structure of human m-calpain.
Despite differences in Ca2+ requirements, the activation mechanism for both calpain 1 and
2 is similar, with binding of multiple Ca2+ ions disrupting the salt bridges that maintain
the cysteine catalytic site (active sites IIa and IIb) in an open conformation to close,
initiating proteolytic activity (Bozoky, et al., 2005). Regulation of calpain activity after
substrate cleavage occurs through autolysis with intermolecular cleavage of domains I
and V resulting in dissociation of subunits. A summary of the diverse members of the
calpain family, their encoding genes and associated polypeptides is shown in Table 1-1.
Page | 7
Table 1-1: Members of the calpain family, encoding genes and associated polypeptides.
(Adapted from Goll, et al., 2003)
The understanding of calpain substrate recognition, specificity, and role in regulatory
modulation of biological processes is crucial and can give valuable information for the
identification of novel calpain substrates and regulatory pathways, a key driver for indepth studies on calpain.
Page | 8
1.3
Calpain and disease implication
1.3.1
Role of calpain in apoptosis
Apoptosis is an essential physiological process, critical in development and tissue
homeostasis. Defective apoptotic processes are known to be implicated in various
diseases. Up or down-regulation of apoptosis may lead to atrophy or to uncontrolled cell
proliferation which results in cancer. Regulation of apoptosis involves a series of signal
molecules, receptors, gene regulating proteins and enzymes. Calpain’s role in the
caspase-cascade signaling system in apoptosis regulation was reviewed by Fan, et al.,
(2005), reporting co-involvement of other molecules such as the inhibitor of apoptosis
protein (IAP), and Bcl-2 family proteins.
Studies have shown that calpains act as both positive and negative regulators in
apoptosis. Chua, et al., (2000) reported negative regulation via consequential inactivation
of caspase-7 and -9 through calpain cleavage. Nakagawa and Yuan (2000) suggested
positive apoptotic regulation through m-calpain cleavage of procaspase-12, forming an
active caspase which cleaves the Bcl-xl loop region, processing an antiapoptotic molecule
to a proapoptotic molecule. Elucidation of calpain’s role in apoptosis is difficult due to
the number of proteolytic enzymes involved in apoptotic pathways, and presence of
common substrates with caspases, e.g. fodrin and ADP-ribosyltransferase/PARP.
Page | 9
1.3.2
Role of calpain in neural degeneration
Dysfunctions in calcium homeostasis may lead to pathological activation of calpain in
several neurodegenerative diseases. Calpain activation via calcium dysregulation leads to
the cleavage of several neuronal substrates involved in neuronal structure and function,
inhibiting neuronal survival mechanisms, leading to acute and chronic neurodegenerative
diseases such as cerebral ischemia, Alzheimer’s disease, Parkinson’s disease and
Huntington’s disease. A comprehensive review of mechanics behind calcium
dysregulation,
calpain-mediated
signaling
mechanism
and
involvement
in
neurodegeneration was reported by Vosler, et al. (2008) and summarized in Figure 1-4.
Page | 10
Figure 1-4: Schematic representation of calpain activation in various neurodegenerative diseases.
Ischemia, traumatic brain injury, and epilepsy cause an acute increase in glutamate release resulting in increased intracellular calcium. Chronic
neurodegenerative diseases AD, ALS, and PRE result in increased NMDA receptor activation, while calcium dysregulation in HD and PD are
attributed to mitochondrial dysfunction. In MS, pathologic calpain activation is initiated by T-cells and propagated by other immune cells such as
macrophages and microglia. (Adapted from Vosler, et al., 2008)
Page | 11
1.4
Challenges in deciphering protease cleavage
In vitro characterization of proteases and substrates involves several biochemical
steps where proteases and protein substrates of interest are purified from biological
origin such as cultured cells and tissues, or in vitro from protein expression studies.
Purification of protein substrates and proteases, to high purity and homogeneity is
challenging, as maintaining native enzymatic activity and structure due to pH and
temperature sensitivity of proteins entails the use of suitable non-denaturing
purification conditions.
Purified protein substrates are incubated with proteases and cleavage products are
analyzed through combinations of gel electrophoresis, reverse phase highperformance liquid chromatography (RP-HPLC), N-terminal sequencing or mass
spectrometry. Alternative approaches may also involve a combination of genetics and
proteomics. Site-directed mutagenesis of genes encoding for a target protein substrate
in animal models to alter amino acid expression at a known cleavage site location to
generate a non-cleavable site constitute a gene knock-out study to examine subsequent
protein activity and function.
With the wide array of biochemical analysis of proteolytic activities in vitro,
knowledge on in vivo activities and relevant substrates of proteases remains unclear.
The key factor being proteases generally do not function individually in vivo, but in
cascades and regulatory circuits, often in the presence of other proteins acting as
substrates, activators, inhibitors and proteases. To overcome this, it is necessary to
examine proteolysis on a system-wide scale, with the collection of proteases
Page | 12
expressed in a cell (protease degradome) and all the substrates of the protease
(substrate degradome) and their state of cleavage in the complex biological
environment.
An example of a system-wide degradomic study was reported by Overall, et al.
(2004), with the development of a dedicated and complete human protease and
inhibitor microarray, CLIP-CHIP, designed for identification of expression levels of
all 715 human proteases, in active homolog and inhibitors in cells and tissues. In the
same study, the development of ICDC (inactive catalytic domain capture); a novel
yeast two-hybrid system to discover protease substrates through capture via a mutated
inactive catalytic domain was also described.
Together, system-wide studies and high-throughput proteomics have enabled the
increasing rate of novel substrates discovery, but not without its limitations.
Identification of proteolytic cleavage products of substrates in its biological
environment may still prove to be a great challenge, highlighting the necessity to
develop complementary tools to aid in the analysis of protease degradomes.
1.5
Project objectives
From numerous studies in recent years, discovery of calpain substrates and growth of
available protein sequence data has led to the creation of useful databases, predictive
algorithms and tools for research applications. Among the recent efforts are CutDB, a
proteolytic events database aimed at documenting in vivo and in vitro for natural
proteins; CaMPDB, a dedicated resource for calpain modulatory proteolysis and
prediction tools; and GPS-CCD, a specialized web-tool developed for the prediction
Page | 13
of calpain cleavage sites. More information on these studies will be discussed in the
later sections of this report.
With increased research detailing the mechanisms of calpain substrate cleavage and
accumulation of data on calpain substrates, the development of computational
prediction methods for calpain substrates is becoming increasingly achievable.
Development of calpain cleavage prediction models can provide screening of a wide
range of novel substrates for potential proteolytic activities in silico, efficiently
assessing their involvement in calpain modulation prior to tedious experimental
procedures to verify positive cleavage or interactions. In addition, calpain’s
involvement in cancers and neurodegenerative diseases potentiates itself as an
important pharmacological drug target for inhibition and therapies. To achieve this,
deeper understanding of the calpain-mediated proteolysis, substrate cleavage site
recognition and specificity has to be addressed. These potential benefits lead to the
main objective of this project to develop an accurate computational model for the
prediction of calpain cleavage sites to serve as a complementary tool to experimental
procedures in the understanding of calpain proteolytic modulation
Page | 14
Chapter 2: Computational approaches to data classification
2.1. Introduction to Support Vector Machine (SVM)
Support vector machines (SVMs) are computational mathematical algorithms used in
supervised learning methods for data and regression analysis and statistical
classification. Training of SVM classifiers are done via positive and negative training
examples. The SVM training algorithm uses the training information to build a model
to predict whether a new data falls into either of the two categories. SVM model
represents the data points as sets of vectors, mapped in high dimensional space,
followed by the construction of one or more hyperplanes to separate the sample data;
1) a separating hyperplane to enable the separation between the distinct classes of data
points, 2) a maximum-margin hyperplane that maximises the margin between the two
categories, 3) a soft margin with user specifiable parameters to control the stringency
of classification of anomalous data points and 4) a kernel function which acts to project
data from a low-dimension space to a higher dimension to improve the classification of
linearly non-separable data (Noble, 2006). The constructed SVM model can be then
used to predict new examples by mapping them in the same space for classification.
Figure 2-1 illustrates the concepts of SVM.
A)
B)
Figure 2-1: Illustration of SVM concepts. A) Two linearly-separable classes, A and B;
represented in two-dimensional space. B) Demonstration of vector mapping from
two-dimensional input space to feature space at higher dimensions using kernel
functions for non-linearly separable data.
Page | 15
2.2. Current perspective in calpain cleavage prediction
Several studies have been done to examine the precise and specific recognition of
cleavage sites of calpains to better understand the mechanisms of modulatory
proteolytic processing. To date, amino acid sequence specificity of cleavage by calpain
has not been established, although some preference with regards to amino acid residues
in the vicinity of calpain cleavage has been reported.
2.2.1.
Sequential determinants of calpain cleavage
In a bid to determine the relationship between structural information and specificity of
substrate recognition by calpain, Tompa, et al. (2004), examined the amino acids
preference of calpain 1 and 2. 49 calpain substrates with 106 sequentially identified
cleavage sites from literature was collected and analyzed for amino acid preference
surrounding the scissile bond. A position specific preference matrix was constructed
from amino acid occurrence in positions P4-P7’ and normalized to the average
frequency of the same amino acid in the entire Swiss-Prot and TrEMBL database.
Preferred residues were reported to be leucine, threonine and valine in the P2 position
and lysine, tyrosine and arginine in the P1 position, coinciding with earlier
comparative specificity and kinetic studies involving naturally occurring peptides and
synthetic fluorogenic substrates with calpain 1 and 2 (Sasaki, et al., 1984) and calpain
activity interference through site-directed mutagenic substitution of amino acids at the
P2 position of αII spectrin (fodrin) cleavage site of Val1175 (Stabach, et al., 1997).
Influence of high order structural elements for calpain cleavage was reported by
Sakai, et al. (1987) from proteolysis of calf thymus histone by calpain 2, notably the
non-cleavage of known susceptible bonds in peptide fragments generated by from
Page | 16
degradation of the intact histones. Contributions of calmodulin (CaM) –binding motif
and vicinity of PEST (Pro, Glu (Asp) and Ser/Thr) regions to calpain substrate
recognition was much debated. Wang, et al. (1989) highlighted the occurrence of
CaM-binding motifs in calpain substrates and cleavage site recognition often occurs
adjacent to a PEST region. Molinari, et al. (1995) however, showed that lower PEST
scores generated by mutation of domains surrounding the CaM-binding regions of
Ca2+-ATPase had no influence on its susceptibility to calpain. These findings, together
with an overriding amino acid preference may be the reason behind the wide array of
calpain substrates and the lack in strong sequence specificity and homology between
reported cleavage sites of different calpain substrates.
2.2.2.
Group-based Prediction System- Calpain Cleavage Detector
(GPS-CCD)
To address the lack of specialized predictors for calpain substrate cleavage sites, GPSCCD (Group-based Prediction System- Calpain Cleavage Detector) was developed by
Liu, et al. (2010) as a web-tool for calpain cleavage sites prediction. GPS-CCD1.0
was based on a previously developed algorithm of GPS2.0 by Xue, et al. (2008),
inferred from the hypothesis that short peptides sharing similar biochemical properties
and 3D structures may be evaluated for similarity via the use of suitable amino acid
substitution matrices. This led to the development of a novel Matrix Mutation (MaM)
approach (Xue, et al. 2008; Ren, et al., 2008, Ren, et al., 2009) which was employed
in the final GPS-CCD1.0. Prediction of a putative calpain cleavage peptide is
accomplished via similarity scoring from pairwise comparison to experimentallyverified cleavage bonds. The first reported GPS-CCD1.0 was developed with 265
experimentally-verified calpain cleavage sites from 102 proteins obtained from data
Page | 17
mining efforts from literature, inclusive of notable contributions by Tompa, et al.
(2004).
Performance evaluation was achieved through leave-one-out validation and 4-, 6-, 8-,
10-fold cross-validations, with best accuracy of 89.80%, sensitivity of 66.42% and
specificity of 89.86%. The current accessible version of GPS-CCD1.0 (dated 26th
February 2011) reported 368 experimentally-verified calpain cleavage sites in 130
proteins. Performance validation of the system achieved best accuracy of 89.98%,
sensitivity of 60.87% and specificity of 90.07%.
2.2.3.
CaMPDB: a resource for calpain modulatory proteolysis
To encapsulate abundant existing information on calpain, its substrates and specific
inhibitor, calpastatin; duVerle, et al. (2010), developed CaMPDB; a resource for
calpain modulatory proteolysis. A total of 267 cleavage sites were collected from 104
known calpain substrates reported in literature. Extensive enhancement of the calpain
database led to the development of three calpain cleavage site prediction tools based
on PSSM, linear and radial basis function (RBF) SVM algorithms. Performance for
the prediction methods were evaluated using Area under the ROC Curve (AUC) with
10x10-fold cross-validation. Maximal values of 69.1%, 77.3% and 80.1% was
reported for the PSSM method (window length, L = 2x30), SVM linear (L = 2x7) and
SVM RBF (L = 2x10) respectively, with best prediction performances of SVM-based
methods achieved within ten amino acids of the cleavage sites. This coincided with
the highly specific and firm binding of calpastatin to the calpain protease domain by
approximately twenty amino acids (Tompa, et al., 2004). Significant increase in
prediction performance with the RBF kernel over the linear kernel suggested strong
Page | 18
non-linear correlations between amino acid positions and cleavage. Window length
variation analysis centered about the cleavage sites revealed asymmetry in the
performance of linear and RBF kernel SVM predictors, with statistically improved
performance on the right side of the cleavage site.
2.3. Summary
We have briefly introduced and discussed the concept behind SVM and the current
perspective to calpain cleavage site prediction, available methodologies and prediction
tools. These tools amongst other calpain studies provide us with an information base to
develop our calpain cleavage site prediction model. For this project, we have chosen to
implement SVM for the development of the calpain cleavage site prediction model due
to its accuracy and performance in classification of biological data, prediction of
protein fold and interactions (Ding and Duchak, 2001; Zhang, et al., 2003), caspase
cleavage (Wee, et al., 2006) and versatility via available kernel functions to aid in
classification of non-linear data. Calpain substrate data collection, processing, SVM
implementation and application of the developed SVM prediction model on a protein
family subset will be discussed in the subsequent chapters.
Page | 19
Chapter 3: Calpain dataset
3.1 Dataset collection
For the development of any computational prediction models, it is critical utilize
accurate and reliable data. Data integrity is of utmost importance in the development of
accurate prediction models. Two distinct problems greatly hinder the development of
computational prediction models: data quality and quantity. Inaccuracy in primary data
during modeling may result in an end model that produces fallacious results whereas
sparse data may affect predictive patterns, leading to significantly less robust
prediction models. To successfully develop the cleavage prediction model for calpain
substrates, it is critical for data to be collected from experimentally-verified calpain
cleaved proteins. To construct the primary calpain dataset, efforts were taken to extract
calpain cleavage information from currently available databases: CutDB, CaMPDB,
GPS-CCD1.0 and through literature searches.
CutDB is a concerted effort by Igarashi, et al. (2007) to document proteolytic events
for natural proteins in vivo or in vitro, organized with three key attributes: protease,
protein substrate and cleavage site information. At publication, the database consisted
of a total of 3,070 proteolytic events for 470 different proteases, with information
captured from publicly available databases, MEROPS, Human Protein Reference
Database (HPRD) and publications. The extraction of all calpain-mediated proteolytic
events deposited in CutDB was achieved via keyword search using “calpain’ carried
out under “Protease definition”. A total of 449 hits were obtained for both calpain 1
and 2 mediated proteolytic events. Events without concise protein sequence or
cleavage information were omitted to obtain a total of 286 calpain cleavage events.
Page | 20
Supplementary calpain substrate data reported by GPS-CCD1.0 was obtained and
referenced for the compilation of the primary calpain dataset. From a total of 265
experimentally-verified calpain cleavage sites entries, 200 cleavage events previously
absent in CutDB was obtained. From CaMPDB, a total of 104 calpain substrates,
labeled “SB” with 267 cleavage sites was reported. From that, 54 calpain substrates
with 120 cleavage events previously absent in CutDB and GPS-CCD1.0 was collected.
To ensure that calpain data collected encompasses all recent publications, a
comprehensive search was conducted on journal articles available in PubMed. Several
permutations of keywords related to calpain substrate cleavage such as “calpain”,
“cleavage” and “substrates” was used as search entries for the period between 1st Jan
2009 through 31st Dec 2010, selected to overlap a minority of existing collected
information. Abstracts of search output were screened for indication of experimental
verification of calpain cleavage events (e.g. in vitro enzymatic assays and cleavage
sites) and suitable publications with available full text were reviewed for exact
cleavage information. Although it may be probable that some journal articles will be
omitted due to the absence of keywords, it is assumed to have minimal impact on the
final dataset. This process resulted in the identification of 7 previously unreported
substrates contributing 20 cleavage sites.
Page | 21
3.2 Data extraction and cleaning
Extraction and cleaning of all collected calpain data was done in four major steps.
Firstly, plausible entries labeled “putative”, “predicted” or “inferred from homology”
were omitted. Secondly, to eliminate typographical errors and ensure consistency of
amino acid residues surrounding the reported scissile bond, the protein sequence and
cleavage site information was cross-referenced to the Uniprot database (Uniprot
Consortium, 2010) through reported Uniprot ID or keyword searches via substrate
name. Ambiguous entries identified were verified with the original publication if
necessary. For example, vimentin, entry SB: 37 in CaMPDB, 10 cleavage sites were
erroneously reported due to single amino acid residue shift to the left due to the
removal of the methionine, “M” initiator by the authors in their amino acid count. All
10 cleavage sites were corrected with reference to original publication and the
canonical vimentin sequence deposited in Uniprot.
Next, full protein sequence of all verified calpain substrates were obtained from
Uniprot for dataset construction. For each reported calpain cleavage site, peptide
sequences of twenty amino acid residues, up and downstream of the reported scissile
bond were extracted, resulting in a set of 40-mer calpain substrate sequences centered
on its reported cleavage site.
Lastly, streamlining of the extracted data was done by the removal of redundant
sequences (100% identity) contributed by high inter-species protein similarity.
Duplicate entries occurring due to protein isoforms were reviewed and condensed
where applicable. Figure 3-1 summarizes the calpain data collection process.
Page | 22
Figure 3-1: A summary of the calpain dataset construction process.
3.3 Summary
A total of 341 unique 40-mer (P20P20’) polypeptide sequences from 130 protein
substrates were collected to constitute the final “cleaned” calpain dataset for the
development of the calpain cleavage prediction model. The final primary datasets of
calpain substrates and their cleavage site information are documented in Appendix A
(Table A-1 and A-2).
Page | 23
Chapter 4: Prediction of Calpain Substrate Cleavage
4.1 Introduction
In calpain cleavage prediction studies discussed earlier, superior performance for
SVM-based prediction methods were reported within ten amino acids flanking the
cleavage sites. Window length variation centered about the cleavage sites hinted
asymmetry in linear and RBF kernel SVM classifier performance with statistically
improved performance on the right of the cleavage site. Analysis of calpain inhibition
by calpastatin, a highly specific inhibitor of calpain, suggested an approximate twenty
amino acid binding specificity of the protease domain. These interesting findings on
calpain substrate cleavage provided the impetus to investigate influences of adjacent
amino acid sequences on calpain substrate cleavage with respect to 1) effects of
varying window length on calpain cleavage site prediction, 2) asymmetrical
contributions of amino acids on calpain substrate binding and cleavage and 3) amino
acid occurrences through linear sequence analysis of the primary calpain dataset.
4.2 Materials and Methods
4.2.1
Calpain datasets
In Chapter 3, we have obtained a calpain dataset containing 341 unique calpain
cleavage sites from 130 substrates. Due to the absence of experimentally determined
calpain non-cleavage sites, random positions were extracted from experimentallyverified calpain substrates. One random non-cleavage site was generated for every
reported cleavage site on the same substrate, resulting in the generation of an equal
number of non-cleavage sites to experimentally-verified calpain cleavage sites. For
each random non-cleavage site, 40-mer peptide sequences were extracted in the same
manner as described earlier. Together, a primary calpain dataset containing 682 entries
Page | 24
of 40-mer peptide sequences centered around its reported cleavage site (341 positive
examples) and non-cleavage site (341 negative examples) was constructed and
designated as the P20P20’ dataset.
4.2.2
Symmetrical subsequence extraction
To investigate the influence of adjacent amino acid sequences on calpain substrate
cleavage, we constructed four additional symmetrical datasets containing the reported
cleavage site flanked by four, eight, twelve and sixteen amino acid residues on either
side, forming varying window lengths, P4P4’, P8P8’, P12P12’ and P16P16’(see Figure 41).
Figure 4-1: Symmetrical subsequence segments extracted for SVM training and testing. For
Human CDK5R2 (Uniprot: Q13319), an extracted sequence window of 40 amino acids is
centered on the octapeptide cleavage site, QQRNRENL (underlined). Amino acids to the left
of the scissile bond (indicated by the inverted triangle) are labeled P1 (N) to P20 (K). Amino
acids to the right of the scissile bond are labeled P1’ (R) to P20’. Curly brackets show the
symmetrical subsequences extracted for SVM implementation, P4P4’, P8P8’, P12P12’, P16P16’
and P20P20’ respectively.
Page | 25
4.2.3
Asymmetrical subsequence extraction
To investigate the hypothesis of asymmetrical contributions of flanking amino acids on
calpain substrate binding and cleavage, we further constructed two asymmetrical
datasets to encapsulate the scissile bond and extension of four and twelve amino acids
on either sides to generate P4P12’ and P12P4’ subsequences respectively (see Figure 42).
Figure 4-2: Asymmetrical subsequence segments extracted for SVM training and testing.
Similar to the previous figure, for Human CDK5R2 (Uniprot: Q13319), an extracted sequence
window of 40 amino acids is centered on the octapeptide cleavage site, QQRNRENL
(underlined). Curly brackets show the asymmetrical subsequences extracted for SVM
implementation, P4P12’ and P12P4’ respectively.
Page | 26
4.2.4
Training and test dataset
Post-extraction of symmetrical and asymmetrical subsequences, the primary calpain
dataset was randomly divided into training and testing datasets and maintained
throughout the subsequent sections of the project.
The training datasets contained 582 sequences (291 positive and negative examples
respectively) and was used for the optimization of SVM parameters and training of the
final SVM classifier for prediction of unseen test examples.
The test dataset contained 100 sequences (50 positive and negative examples
respectively). The test dataset was used for the performance evaluation of the final
classifier. Figure 4-3 shows the segregation of various symmetrical and asymmetrical
datasets.
Page | 27
Figure 4-3: A schematic representation of datasets used for SVM training and testing. The
primary 40-mer (P20P20’) dataset consist of non-redundant calpain cleavage sites (positive
examples) and an equal number of non cleavage sites (negative examples). The P20P20’ dataset
constitutes the parent sequence for the derivation of the symmetrical P4P4’, P8P8’, P12P12’ and
P16P16’ and asymmetrical P4P12’ and P12P4’ subsequences respectively.
4.2.5
Vector encoding schemes
To encapsulate the extracted sequence information into a SVM-compatible format for
training and testing, the sequences were transformed into input vectors in simple binary
and bi-profile manner using Bayes Feature Extraction (BFE) encoding schemes.
Page | 28
4.2.5.1 Simple binary encoding
In simple binary encoding, sequences were transformed into n-dimensional vectors
using an orthonormal encoding scheme, with each amino acid represented by a 20dimensional vector, composed of either zero or one as elements.
For example, alanine was represented as [1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0] and
cysteine as [0,1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0]. For the P20P20’ dataset, each
sequence was represented by an 800-dimensional vector. Symmetrical sequences in the
P4P4’, P8P8’, P12P12’ and P16P16’ datasets were represented by 160, 320, 480 and 640dimensional vectors respectively. Asymmetrical P4P12’ and P12P4’ subsequences were
both represented by 320-dimensional vectors.
4.2.5.2 Bayes Feature Extraction (BFE) encoding
Key concepts on bi-profile vector encoding was reported by Shao, et al. (2009) in their
novel approach in computational identification of post-translational protein
methylation sites through bi-profile Bayes Feature Extraction combined with support
vector machines. In BFE, feature vectors are encoded in a bi-profile manner containing
attributes from positive and negative position-specific profiles. Profiles were generated
through the calculation of the frequency of occurrence of each amino acid at each
position of the extracted peptide sequence in the experimentally-verified calpain
cleavage sites (positive) and randomly generated calpain non-cleavage sites (negative)
respectively. For BFE, a 40-mer input peptide will be encoded by an 80-dimensional
(40 x 2) feature vector containing residue information from both positive and negative
spaces.
Page | 29
4.2.6
SVM implementation
For SVM implementation, we employed the freely downloadable LIBSVM package
developed by Chang and Lin (2001). SVM is based on the structural minimization
principle from statistical learning theory. A set of positive and negative examples can
be represented by feature vectors xi (i = 1, 2,…, N) with corresponding class labels yi
∈{+1,−1}. SVM classifier training involves the mapping of input examples onto a
high dimensional space, aided by the use of a kernel function, followed by the
definition of a separating hyperplane that differentiates the two classes with maximal
margin and minimal error. The resulting decision function for predictions of unseen
examples is given as:
Where
represents the kernel function and parameters are determined by
maximizing the following:
Under the conditions,
Variable C serves as the regularization parameter controlling balance of margin and
classification error. Based on previous findings of non-linearity between amino acid
Page | 30
positions and cleavage and superior performance of RBF kernel-based SVM classifiers
in CaMPDB, we have chosen to implement the RBF kernel given by:
4.2.7
SVM optimization
Implementation of the RBF kernel-based SVM classifiers necessitates the optimization
of two parameters; γ, the RBF kernel capacity determinant and the regularization
parameter, C. To optimize the SVM parameters γ and C, 10-fold cross-validation was
applied on each of the training datasets via grid search with SVM parameters stepped
through combinations of 0.001, 0.01, 0.1, 1, 10 and 100 for both γ and C. During 10fold cross-validation, the input training dataset was divided into 10 subsets, where 9
subsets were used for the training of the classifier followed by testing with the
remaining subset. The process is repeated 10 times, each with a different subset for
testing, ensuring all subsets are used for both training and testing. 10-fold crossvalidation accuracy scores generated were collected and tabulated in grid search tables
for each individual subsequence datasets in both BFE and simple binary encoding.
Figure 4-4 shows the general workflow for SVM development. Grid search
optimization tables generated are documented in Appendix B.
Page | 31
Figure 4-4: Flowchart of SVM workflow. (a)Primary dataset, (b)Training dataset, (c)Test
dataset, (d) 10-fold cross-validation, (e) Obtaining final accuracy (each C and γ pair
following grid-search method), (f) Collection of training and validation to obtain optimal C
and γ values, (g) Retraining of SVM model with optimized C and γ value before proceeding
testing with designated test dataset (c).
Page | 32
4.2.8
SVM training and testing
From the grid search optimization of individual subsequence datasets, optimal values
of γ and C obtained were used for the training the SVM classifiers. Final trained SVM
classifiers were used to predict the test datasets. SVM performance and effectiveness in
predicting calpain cleavage sites was measured by the calculation of the following
quantitative variables:
i. TP, true positives – the number of correctly classified cleavage sites
ii. FP, false positives – the number of incorrectly classified non-cleavage sites
iii. TN, true negatives – the number of correctly classified non-cleavage sites
iv. FN, false negatives – the number of incorrectly classified cleavage sites
From the variables above, statistical metrics of Sensitivity (Sn) and Specificity (Sp)
were computed to evaluate the ability of the prediction model to correctly classify
calpain cleavage or non-cleavage sites respectively.
Overall prediction model performance was assessed by computing Accuracy (ACC):
One major drawback of the above metrics is that a threshold must be chosen to
distinguish between predicted positives and negatives. During comparison of two
prediction methods, differences in sensitivity and specificity may be a result of
Page | 33
thresholds parameters, in actual scenario, the two methods may be identical should
threshold adjustment be made on one of the methods.
To avoid these instances, calculation of the area under the receiver operator
characteristics curve (AROC) was also applied as a non-parametric measure of
predictive performance. The ROC curve is constructed by using different values of the
threshold to plot the true positive proportion (TPP) against the false-positive proportion
(FPP), given by:
To generate the predictive scores of test datasets for AROC calculation, SVMlight
(Vapnik, 1995; Joachims, 1999) was implemented and trained with all subsequence
datasets with optimized γ and C previously obtained from grid search optimization in
LIBSVM. Prediction results from each test dataset were checked for consistency to
LIBSVM classification results and used as input to ROC analysis, a web-based
calculator for ROC curves (Eng, 2006). AROC value close to 0 is indicative of negative
correlation and at 0.5, no correlation. AROC values greater than 0.7 indicates a useful
prediction performance and values above 0.85 indicates good prediction performance.
Performance metrics: Sensitivity (Sn), Specificity (Sp), Accuracy (ACC) and AROC
values generated were tabulated. Combined ROC curves are documented under
Appendix C.
Page | 34
4.2.9
Linear sequence analysis of primary calpain dataset
4.2.9.1 Relative position-specific amino acid propensity
The relative position-specific amino acid propensity, Px, of an amino acid is a
quantitative indicator of the probability of the amino acid existing at a specific
location on a protein sequence. Individual position-specific amino acid intensities in
the positive and negative datasets were derived for the primary P20P20’dataset
containing 40-mer sequences by:
(1) Position-specific amino acid intensities for positive dataset: Number of amino
acid X at position I in the positive dataset/Total of number of sequences in
positive dataset.
(2) Position-specific amino acid intensities for negative dataset: Number of amino
acid X at position I in the negative dataset/Total of number of sequences in
negative dataset.
Propensity, defined as the ratio of the frequency of the occurrence of an amino acid in
experimentally-verified calpain cleaved substrate sequence population (positive
examples) to the frequency of the occurrence of the same amino acid in the random
non-cleaved substrate sequence population (negative examples) at a specific position
was derived by:
(3)
Relative position-specific amino acid propensity, Px = (1)/ (2)
For visualization of positive, negative and calculated propensity, three heatmaps were
generated using above calculated values. Respective 20 x 40 matrices were
constructed for heatmap generation using R programming (R Development Core
Team, 2010).
Page | 35
4.2.9.2 Sequence logo representation of calpain cleavage events
Sequence logos were developed with the aim to display and analyze patterns in
sequence conservation (Schneider and Stephens, 1990). To further visualize positionspecific amino acid occurrence and patterns in sequence conservation surrounding
reported experimentally-verified calpain cleavage sites, the positive P20P20’dataset was
used as input for the generation of a sequence logo through multiple sequence
alignment using WebLogo, developed by Crooks, et al. (2004).
Protein logos generated from input sequences enables graphical representations of
patterns and description of sequence similarity to reveal significant features of the
alignment such as amino acid conservation that could be of importance in substrate
recognition by calpain, which can be difficult to visualize in linear sequence data.
Page | 36
4.3 Results and discussion
4.3.1
Performance metrics of SVM prediction
Table 4-1: Summary of SVM prediction performance of classifiers trained using various
subsequences and encoding strategies.
Page | 37
Table 4-1 summarizes the optimized γ and C values and performance metrics of
Sensitivity (Sn); Specificity (Sp); Accuracy (ACC) and AROC values generated for all
final SVM classifiers using various subsequence datasets.
Simple binary and BFE encoded schemes for symmetrical subsequences are
represented by SVM-P4P4’ to SVM-P20P20’ and Bayes-SVM-P4P4’ to Bayes-SVMP20P20’ respectively. Asymmetrical encoded schemes are represented with SVMP4P12’, SVM-P12P4’ and Bayes-SVM-P4P12’, Bayes-SVM-P12P4’ respectively.
For symmetrical simple binary encoded schemes, maximal performance of was
observed for the SVM-P4P4’ classifier at accuracy of 77%, with sensitivity of 70%,
specificity of 84% and AROC of 0.832. Overall performance of symmetrical simple
binary encoded schemes was fairly consistent with accuracy ranging from 71 to 77%
and sensitivity and specificity between 62 to 74% and 76 to 84% respectively.
Analysis of AROC values indicated useful prediction performance, with values in the
range of 0.789 to 0.834.
The use of BFE schemes significantly improved performance across all symmetrical
subsequence windows. Performance metrics, accuracy and AROC scores for each
subsequence window were consistently higher than those obtained from classifiers
trained with simple binary encoded schemes. The best BFE classifier (Bayes-SVMP20P20’) achieved an accuracy of 85%, sensitivity of 86%, specificity of 84% and AROC
of 0.927. Graphical representation of the trends in SVM performance (accuracy and
AROC scores) across various subsequence windows are shown in Figure 4-5.
Page | 38
Figure 4-5: Graphical representation of the trends in SVM classifiers performance in terms of
A) accuracy and B) AROC scores for various subsequence windows.
Page | 39
Interestingly, two differing trends relating to subsequence window lengths were
observed between prediction performance of simple binary and BFE encoded
schemes. For BFE encoded schemes, a gradual increase in accuracy and AROC was
observed as the window length of peptide subsequence increases. In contrast,
performance metrics in the simple binary encoded scheme was highest at the classifier
trained with the shortest subsequence, SVM-P4P4’, and decreases till saturation at
SVM-P12P12’ and increases slightly with the increase of subsequence window length
to 32 and 40-mer (SVM-P16P16’ and SVM-P20P20’). This observation is
uncharacteristic as longer subsequence windows generally allow the encapsulation of
more information or features surrounding the P1-P1’ scissile bond which aid in
prediction.
Similar observations were noted in simple binary and BFE encoded schemes for
asymmetrical subsequences are represented by SVM-P4P12’ and SVM-P12P4’, designed
to investigate the hypothesis of asymmetrical contributions of flanking amino acids on
calpain substrate binding and cleavage previously reported by duVerle, et al. (2010).
For the asymmetrical simple binary encoded schemes, slight improvement in accuracy
was observed with the SVM-P4P12’ classifier at 75%, compared to the SVM-P12P4’
classifier which achieved 72%. The right-primed classifier SVM-P4P12’ also indicated
better performance in the differentiation of calpain non-cleavage sites with specificity
of 82% compared to 72% obtained in SVM-P12P4’, this however, with slight decreased
sensitivity at 68%. An increase in prediction performance was observed in the SVMP12P4’ classifier (AROC 0.795) when compared to the SVM-P4P12’ classifier (AROC
0.788).
Page | 40
Consistent to symmetrical subsequence trained SVM classifiers, an overall
improvement in prediction performance was evident when BFE was employed on
asymmetrical subsequences. SVM-P4P12’ obtained an accuracy of 82% (sensitivity of
78%, specificity of 86%) and AROC of 0.866 and SVM-P12P4’ obtained an accuracy of
81% (sensitivity of 84%, specificity of 78%) and AROC of 0.905. Observations
discussed in asymmetrical simple binary encoded schemes were also prevalent.
Based on our findings, there is no strong indication of preferential asymmetrical amino
acid sequences extension on either side of experimentally-verified calpain cleavage
sites contributing to substrate cleavage. However, plausibility of asymmetrical amino
acids contributions to calpain substrate recognition and cleavage should not be ruled
out without further investigation. Further directions will be discussed in later sections
of this report.
In the comparison of our SVM implementation to previous studies reporting best
prediction performance of accuracy 89.98%, sensitivity of 60.87% and specificity of
90.07% by GPS-CCD1.0 and maximal AROC values of 69.1% for PSSM, 77.3% for
SVM linear classifier and 80.1% for SVM RBF classifier by CaMPDB, we can infer
comparable, if not, superior performance in calpain substrate cleavage prediction
using BFE encoded schemes with symmetrical subsequences to existing methods.
Predictive performance of each method however, may be subjective. Reported
prediction performance for both GPS-CCD1.0 and CaMPDB were generated from
cross-validation, in the absence of independent out-of-sample testing. Although
authors from GPS-CCD1.0 reported the prediction of calpain cleavage sites in several
Page | 41
proteins such as caspase-14 (9 sites) and dog interleukin-1 alpha (6 sites), previously
experimentally-identified to be cleaved by calpains but without exact cleavage sites
reported. These predictions are not indicative of prediction accuracy due to absence
of experimental verification. In addition, the quantity and accuracy of calpain
substrate data used (265 and 368 entries in CaMPDB and GPS-CCD1.0 respectively)
in both studies may also affect prediction performance as no detailed procedure was
documented for data cleaning and verification in both mentioned studies.
4.3.2
Relative position-specific amino acid propensity
Figure 4-6 shows the heatmaps generated using amino acid intensities in the 40-mer
positive and negative examples and calculated propensity, Px. From the heatmap of the
positive dataset, enrichment of several amino acids at around the calpain cleavage site
is observed, especially at positions P2 to P3’. Leucine (L) is enriched at positions P2
(0.317) and P2’ (0.114). Serine (S) was found to occur frequently at position P1 and P1’
at 0.129 and 0.194. Positions P3’ and P4’ showed elevated proline (P) occurrence, at
0.220 and 0.123 respectively. Amino acid of different properties, alanine (A,
hydrophobic), glutamic acid (E, acidic), glycine and serine (G and S, polar) were found
to occur at moderate levels throughout the positive dataset.
From the heatmap of the negative dataset, enrichment of leucine (L), glutamic acid (E),
threonine (T), alanine (A), glycine (G) and lysine (K) was observed throughout the
length of the randomly generated 40-mer dataset. In both positive and negative
examples, cysteine (C), histidine (H), methionine (M) and tryptophan (W) residues
were the least occurring amino acids in the both the 40-mer positive and negative
dataset.
Page | 42
Calculated propensity Px, given by the ratio of position-specific amino acid propensity
between the positive and negative dataset, allows visualization of amino acid
differentiation in respective positions. Positions with high Px values indicates a high
likelihood of an amino acid occurring at the location compared to that in the negative
examples, vice versa with small Px values. From the heatmap and calculated average Px
values, significant propensity of alanine, methionine, proline, serine and tryptophan
residues was observed in some regions surrounding the cleavage site, in particular the
P4-P4’ segments and downstream regions. Leucine enrichment was distinct at position
P2 at 3.00, due to its significantly higher intensity in the positive dataset (0.317) despite
of its high occurrence in the negative dataset (0.106). Amino acid intensity matrices,
calculated and average Px of the 40-mer sequences are documented in Appendix D.
Page | 43
A)
B)
C)
Figure 4-6: Heatmaps of position-specific amino acid intensities of A) positive
examples, B) negative examples and C) calculated propensity Px. Vertical axis
contains the range of twenty amino acids, while the horizontal axis represents each
residue position of the 40-mer input sequences. Increasing color intensities in each
heatmap (blue for positive examples and propensity, Px, and red for negative
examples respectively) indicate position-specific amino acid enrichment.
Page | 44
4.3.3
Sequence logo representation of calpain cleavage events
Figure 4-7: Sequence logo representation of experimentally-verified calpain cleavage
events. A) Logo of 40-mer sequences (P20P20’) centered on the experimentally-verified
calpain cleavage sites. B) Expanded view of sequence logo showing P8P8’subsequence
segment.
Page | 45
Logos generated consists of one stack of letters representing each position of the input
sequence. Sequence conservation is indicated by the overall height of each stack,
measured in bits. Relative frequency of corresponding amino acids is indicated by the
height of symbols within the stack. Amino acids are represented by colors according to
their chemical properties; polar amino acids (G, S, T, Y, C, Q, N) labeled green, basic
(K, R, H) labeled blue; acidic (D and E) red and hydrophobic (A, V, L, I, P, W, F, M)
labeled black. Position-specific sequence conservation, Rseq, is defined as the
difference between the maximum possible entropy (Smax) and the entropy of the
observed symbol distribution (Sobs):
Pn: observed position-specific frequency of symbol n; N: Number of sequence-specific
symbols, equivalent to 20 for proteins. Maximum sequence conservation per site is
given by log220, approximately 4.32 bits for proteins sequences.
From Figure 4-7, there is no strong evidence of sequence conservation throughout the
40-mer input, with a wide range of amino acid residues occurring around the reported
calpain cleavage site. Position-specific amino acid conservation was observed to fall
below 0.5 bits, with the exception for the pentapeptide P2-P3’, with maxima at
approximately 0.75 bits. A comparison of amino acid prevalence from the sequence
logo generated to findings by Tompa et al. (2004) is compiled in Table 4-2.
Page | 46
Table 4-2: Comparison of position-specific amino acid prevalence in the generated 40-mer
sequence logo versus findings by Tompa, et al. (2004).
Amino acid prevalence at P2 was observed to be leucine, and valine, threonine at lower
levels, consistent with the Tompa study. Slight differences were observed at other
positions in our dataset; with position P1 occurrence of serine and glycine versus
lysine, tyrosine and arginine; position P1’ with serine, alanine and leucine versus
serine, threonine and alanine; positions P2’ (leucine, glutamic acid), P3’ (proline,
lysine, alanine) and P4’ (proline, serine, glutamic acid) versus a significant proline
prevalence reported in the Tompa study. A key contribution for this difference could
be the discovery of a more diverse range of calpain substrates since the study
conducted, where a limited collection of 106 cleavage sites from 49 calpain substrates
was analyzed. Another correlation to a previous study by Wang et al. discussed earlier
reporting the influence of PEST regions to calpain cleavage site recognition can be
observed from the low level conservation of PEST sequence motifs between P8-P1and
P3’-P8’.
Page | 47
These observations of diverse amino acid preference and cleavage by calpain is not
unexpected with such variability arising from calpain’s ability to proteolyze a wide
array of substrates in vivo and in vitro, involved various cellular processes. Possible
evolution of substrate binding sites in calpains for recognition of a wide range of
amino acid sequences in contrast to strong binding to highly specific and conserved
amino acid residues surrounding the cleavage site, distinguishes itself from other
cysteine proteases such as caspases which exhibit specificity for substrate cleavage
after an aspartic acid residue (D) at P1. These factors lead to much difficulty in the
elucidation of calpain substrate cleavage mechanisms up till today.
Page | 48
Chapter 5: Prediction of Receptor Tyrosine Kinases (RTKs) Family
Proteins
5.1 Introduction to Receptor Tyrosine Kinases (RTKs)
Protein kinases are key enzymes involved in numerous biological regulatory roles
through the protein function modification via catalytic transfer of phosphate groups
from ATP (adenosine triphosphate) molecules to specific amino acids on proteins
(phosphorylation). Phosphorlyation is an important form of post-translational protein
modification which results in functional changes in the target protein, with regards to
enzyme activity, location and protein association. Based on amino acid specificity,
protein kinases are classified into protein serine/threonine or tyrosine kinases. More
importantly, protein kinases regulate a wide variety of cellular functions including
cytoskeletal rearrangements and differentiation, cellular growth and apoptosis and
signal transduction.
A kinome study to catalogue the protein kinases in the human genome by Manning, et
al. (2002), discovered more than 500 genes encoding protein kinases, approximately
2% of all human genes. 385 were identified to be serine/threonine specific, 90 being
tyrosine specific and 43 being tyrosine kinase-like proteins. Among protein tyrosine
kinases, a large portion belonged to receptor tyrosine kinases (RTKs). The RTK
family of proteins includes approximately 20 classes, including epidermal growth
factor receptor (EGF), hepatocyte growth factor receptor (HGF), leukocyte tyrosine
receptor kinase, (LTK); RET proto-oncogene receptor (RET) and vascular endothelial
growth factor receptors (VEGF), amongst many others.
Page | 49
Hubbard and Miller (2007) reviewed RTKs as single-pass, type I transmembrane
receptors, important agents of signal transduction pathways. RTKs are generally
activated through ligand-induced oligomerization, often dimerization, bringing the
cytoplasmic tyrosine kinase domains together in close proximity, facilitating
autophosphorylation in trans of tyrosine residues in the kinase activation loop or
juxtamembrane region, inducing conformational changes that stabilize the active state
of the kinase. These phosphorylated tyrosine residues serve as binding sites for
downstream signaling or adapter proteins, and initiate subsequent cellular responses
through various signal transduction pathways. As essential components to cellular
signaling pathways and participation as growth factor receptors, mutation and
structural aberrations in RTKs are often implicated in onset and progression of
cancers.
Wee, et al. (2009) tested the hypothesis of RTK protein family regulation via caspase
proteolysis due to their common implication in apoptosis. Caspases are recognized as
the main group of enzymes involved in apoptosis, with sequential activation of a
hierarchy of caspases after death receptor stimulation in apoptotic cells. Due to
overlapping substrate specificities and evidence of caspase regulation by calpains
discussed earlier, there is increased interest to examine the possibility of calpain
involvement in RTK regulation, through direct proteolytic modulation or as factors to
the apoptosis cascade.
Page | 50
5.2 Prediction of calpain cleavage of RTKs
To examine the efficacy of calpain cleavage prediction on a protein family, we
applied the best performing SVM classifier (Bayes-SVM-P20P20’) to predict potential
cleavage sites on a subset of the RTK family: EGF receptors (EGFR and Erbb2), HGF
receptor (MET), LTK receptor (ALK) and RET receptor (RET).
Full protein sequences for EGFR (P00533), Erbb2 (P04626), MET (P08581), ALK
(Q9UM73) and RET (P07949) were collected from the Uniprot database. 40-mer
subsequences were extracted via single amino acid increment moving windows for
interrogation of each amino acid as potential calpain cleavage sites, with the exception
of residues 1-19 and 19 residues upstream of the last residue of the full protein
sequence (see Figure 5-1). The BFE encoding scheme was employed on the extracted
subsequences described in earlier sections and predicted using Bayes-SVM-P20P20’
classifiers implemented in both LIBSVM and SVMlight.
Figure 5-1: Construction of the 40-mer moving window in EGFR (P00533).
Labels 1 and 1210 refer to first and last residue of the EGFR protein. The red asterisks
between P-A (20th residue) and S-T (1190th residue) represent the first and last
interrogated cleavage sites. The light blue, red and green boxes highlight the 1st, 2nd
and 3rd extracted 40-mer subsequence, the purple and blue boxes indicate the 1170 and
1171st subsequences.
Page | 51
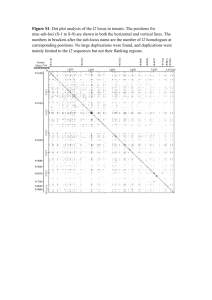
Table 5-1 shows the schematic maps of predicted calpain cleavage sites on the RTK
family subset, with prediction scores ≥ 1.0 in SVMlight. All members were predicted to
possess calpain cleavage sites with distribution, in most cases, throughout extra- and
intracellular regions. All selected kinases, with the exception of ALK, had predicted
calpain cleavage sites on the tyrosone kinase domain. These domains serve as
important mediators of signal transduction for RTKs and structural alterations may
lead to aberration in downstream signal transduction. EGFR, Erbb2 and MET were
predicted to possess calpain cleavage sites proximal to the membrane on the
cytoplasmic side of the receptor, suggesting the formation of an intracellular fragment
and a membrane-bound region. This may lead to possible implications in downstream
signaling, especially to normal RTK signaling pathways from competitive binding of
ligands between intact receptors and cleavage by-products of membrane-bound
receptors. With the numerous possible permutations of proteolytic fragments generated
from extracellular, intracellular and kinase domain cleavage, their involvement in
downstream functional implications such as anti or pro-apoptotic activity may prove
worthy of further experimental investigations.
5.3 Summary
In summary, a prediction of calpain cleavage sites on a protein subset of the RTK
family was conducted, with results suggesting possible calpain regulation of RTK
activity. Considering calpain’s involvement in pro and anti-apoptotic regulation via
cleavage of caspases, a likelihood of calpain being a factor in caspase-mediated RTK
regulation, RTK signaling and the production of pro-apoptotic intracellular fragments
may also be hypothesized. These hypotheses necessitate further in-depth biochemical
and structural studies on calpain mediated RTK cleavage for validation.
Page | 52
Table 5-1: Schematic maps of predicted calpain cleavage sites on the receptor tyrosine kinase (RTK) family subset. P1 positions of
predicted cleavage sites on each RTK family subset proteins are listed. Grey sections indicates location of cleavage site within the
extracellular domain, green indicates location within transmembrane domain, light blue indicates location within intracellular domain
Page | 53
and darker blue indicates location within kinase domain.
Chapter 6: Conclusion
6.1 Summary of project report
Calpains constitute an important family of intracellular, Ca2+-dependent, nonlysosomal cysteine proteases which exhibits limited proteolysis of its substrates at
neutral pH. Through cleavage of a diverse range of substrates, calpains are known to
modulate a wide range of biological processes such as apoptosis, cytoskeletal
organization and neuroendocrine secretory pathways. With calpain involvement in
diseases such as cancers and neurodegenerative diseases, it is wise to consider calpains
as clinically important targets for inhibition and therapy development, highlighting the
necessity to characterize the calpain degradome.
To date, the mechanisms of substrate recognition and cleavage by calpain have not
been fully established. However, with increasing amount of research efforts aimed at
unraveling calpain modulatory mechanisms; increasing amounts of data on calpain
substrate is becoming available. This increases the feasibility of developing calpain
cleavage prediction models to screen novel substrates for potential cleavage activities
in silico, allowing protein substrate studies to be efficiently assessed prior to tedious
experimental procedures. Recent reports of computational methods in calpain cleavage
sites prediction have been successful to certain extents, and in the midst, revealed
interesting observations to calpain cleavage mechanisms with regards to amino acid
sequences conservation and asymmetrical contributions of amino acids to calpain
recognition of substrates. These promising results provided the impetus to implement
an SVM-based method to investigate the efficacy of developing a calpain substrate
cleavage prediction tool to complement experimental procedures.
Page | 54
In our study, a total of 341 unique calpain substrate cleavage sites from 130
experimentally-verified substrates were obtained from available databases and
literature to form the foundation for SVM prediction model development. To widen
our scope of investigation, a combined approach of linear sequence analysis of amino
acid conservation and propensity; symmetrical and asymmetrical subsequence
extraction together with simple binary and bi-profile BFE encoding strategies were
employed. Sequence analysis via sequence logo and heatmap generation as well as
derivation of amino acid propensity revealed correlation to previous sequential studies
by Tompa, et al. (2004) and also significant propensity for alanine, tryptophan,
methionine, proline and serine residues within the P4-P4’ window and downstream
regions of cleavage sites. Our best SVM-based calpain cleavage site prediction model
(Bayes-SVM-P20P20’) achieved an accuracy of 85%, sensitivity of 86%, specificity of
84% and AROC of 0.927, comparable to existing published methods.
To explore the efficacy of our SVM-based prediction model to elucidate the calpain
degradome, we applied our best performing prediction model on subset of the RTKs
family, to predict for potential calpain substrates. All tested members were predicted to
possess calpain cleavage sites distributed throughout extra- and intracellular regions.
RTKs belong to a class of membrane receptors, with critical roles in cellular signaling
pathways and growth. Mutation and structural aberrations in RTKs are often
implicated in onset and progression of cancers. Prediction results suggested possible
regulation of RTK activity by calpains. With overlapping substrate specificities and coinvolvement in apoptosis between calpain and caspases, there is a likelihood of calpain
being a factor in caspase mediated RTK regulation leading to termination or
impairment of RTK signaling and initiation of apoptosis.
Page | 55
6.2 Recommendations and future direction
From our study, the efficacy of developing a computational model for the in silico
prediction of calpain substrates as a complementary tool to experimental procedures
in the understanding the role of its proteolytic modulation is highly feasible, with
developed prediction models showed promising performance when compared to
existing published methods. A logical question arises next on how prediction accuracy
may be improved. A wide array of approaches may be tested to improve the
performance of calpain prediction; however, most approaches will be extremely time
and computationally intensive and may prove challenging for the short timeframe of
the Capstone project.
Firstly, the calpain substrate database collection of cleavage events may be expanded
with deeper investigation on recently published articles. Expert curation methods can
also be employed to enhance the database such as the classification of substrates by
the calpain implicated for cleavage, e.g. calpain 1 or 2; which may be helpful in
determining similarities or differences between substrate populations. Studies on
sequence similarity may be used to investigate the occurrence of highly conserved or
repetitive sequences. In our study, only the removal of redundant occurrences that are
100% identical was employed. Development of prediction models with streamlined
calpain datasets (e.g. with removal of 95% similar sequences) may be favorable for
training of the SVM classifier, possibly removing noise sequences which may affect
prediction. Also, the calpain substrate database and developed models may be
implemented on dedicated servers and made available publicly for academic usage.
Page | 56
As discussed earlier, proteases do not function alone in biological systems but in
cascades and regulatory circuits, together with other various proteins. In addition,
majority of current prediction tools and methods are developed based on linear
peptide sequences or peptide libraries and cleavage sites predicted are only indicative
of consensus sequences cleaved by calpains but may not be cleaved in reality. Factors
influencing substrate recognition, binding and cleavage by calpains in vitro and in
vivo should be taken into consideration. Structural information such as protein folding
and secondary structure (α-helix and β-sheets), hydrophobicity, post-translational
modification information and solvent accessibility studies may aid in providing clues
to structure-function relationships. More in-depth and comprehensive investigation of
asymmetrical contributions of flanking amino acid residues to calpain cleavage may
also be achieved through the creation of asymmetrical moving windows to examine
contributions at single amino acid residue levels and how it affects prediction
performance.
In summary, with continual advancement in experimental identification of calpain
substrates together with improved methodologies on information usage and
exploration of suitable features for data analysis, the capabilities of machine learning
techniques such as SVM can be improved and maximized.
Page | 57
Part 2
Chapter 7: Critical reviews and reflections
As a working adult, it is unavoidable to encounter time constraints, handling both
work and heavy workloads from the project together with ongoing modules. During
the Capstone project, time management has proven to be critical to ensure completion
within the deadline. A sense of work prioritization and frequent consultation with my
project supervisor allowed me to pinpoint critical tasks to be completed in due time
and keep within the scope of the initial objectives set during project commencement,
even with new ideas arising from the abundant articles related to calpain research.
From the skills review and project plan drafting during proposal writing, personal
strengths and weakness assessment was helpful in judging which areas and tasks
required additional time and effort. Previous experience and knowledge in proteomics
enabled a much needed head start, being able to understand journal articles and
publications on protein studies and information such as protein databases and tools.
Much emphasis for the initial stage of the project was on the collection of
experimentally-verified calpain substrate cleavage sites and verification of the
collected information to ensure data integrity and consistency. This procedure proved
to be tedious and complicated due to variations in calpain information from available
sources, and the necessity of manual curation of collected information to Uniprot to
ensure data accuracy. The Gantt chart detailing the project plan is documented in
Appendix E.
A key weakness identified in early project planning was the lack of strong knowledge
in programming languages. This weakness led to time spent on experimentation with
Page | 58
Microsoft Excel for data processing. With trial and error and guidance from my
project supervisor, valuable knowledge was gained in Microsoft Excel functions and
data manipulation techniques widely used in this project the process of data cleaning,
subsequence extractions, random negative example extraction, simple binary and
Bayes feature encoding.
During results analysis of the various generated SVM prediction models, another
hurdle was encountered. Firstly, to generate linear sequential analysis of our collected
calpain dataset, heatmaps were required for clearer graphical representations.
Although it was possible to generate heatmaps using Microsoft Excel, gradient color
palettes were limited for classification of the calculated amino acid intensities. To
resolve this problem, knowledge in the R programming software environment for
statistical computing and graphics had to be acquired from scratch and employed.
Another setback encountered during results analysis was the computation of AROC
values as a measure of prediction performance. Although a LIBSVM tool for the
analysis of ROC curve was available in Python language interface, it required the
installation of Gnuplot, a command-line driven graphing utility, and Microsoft Visual
Studio. Trials with this setup failed to generate the required ROC curve. With the
guidance of my project supervisor, I was able to swiftly switch to investigate the
implementation of SVMlight to generate prediction scores as input to online tools for
ROC generation developed by John Hopkins University, avoiding time loss from
figuring out an unfamiliar programming language.
Page | 59
In summary, the undertaking of the Capstone project has been a very rewarding
experience, both academically and on a personal level. Valuable knowledge was
gained in current and growing repertoire of bioinformatics, especially in machine
learning applications and development of personal skills such as time management,
methodological and critical thinking and attention to details.
Page | 60
REFERENCES
Berti, P.J., and Storer, A.C. (1995). Alignment/phylogeny of the papain superfamily
of cysteine proteases. J. Mol. Biol. 246, 273-283.
Bozoky, Z., Alexa, A., Tompa, P., and Friedrich, P. (2005). Multiple interactions of
the ‘transducer’ govern its function in calpain activation by Ca2+. J. Biochem. 388,
741–744.
Chang, C.C., and Lin, C.J. (2001). LIBSVM: a library for support vector machines.
Retrieved from: http://www.csie.ntu.edu.tw/~cjlin/libsvm
Chua, B.T., Guo, K., and Li, P. (2000). Direct cleavage by the calcium-activated
protease calpain can lead to inactivation of caspases. J Biol Chem. 275, 5131–
5135.
Crooks, G.E., Hon, G., Chandonia, J.M., and Brenner, S.E. (2004). WebLogo: a
sequence logo generator. Genome Res. 14(6), 1188-1190.
Dear, N., Matena, K., Vingron, M., and Boehm, T. (1997). A new subfamily of
vertebrate calpains lacking a calmodulin-like domain: implications for calpain
regulation and evolution. Genomics. 45, 175–184.
Ding, C.H.Q., and Dubchak, I. (2001). Multi-class protein fold recognition using
support vector machines and neural networks. Bioinformatics. 17, 349-358.
duVerle, D., Takigawa, I., Ono, Y., Sorimachi, H., and Mamitsuka, H. (2010).
CaMPDB: a resource for calpain and modulatory proteolysis. Genome Inform. 22,
202-213.
Eng, J. (n.d.). ROC analysis: web-based calculator for ROC curves. Retrieved June 1,
2010, from http://www.jrocfit.org
Evans, J. S., and Turner, M. D. (2007). Emerging functions of the calpain superfamily
of cysteine proteases in neuroendocrine secretory pathways. J. Neurochem. 103,
849-859.
Fan, T.J., Han, L.H., Cong, R.S. and Liang, J. (2005). Caspase family proteases and
apoptosis. Acta Biochim Biophys Sin (Shanghai). 37(11), 719-727.
Franco, S. J., and Huttenlocher, A. (2005). Regulating cell migration: calpains make
the cut. Cell Sci. 118, 3829–3838.
Franz, T., Vingron, M., Boehm, T., and Dear, T. N. (1999). Capn7: a highly divergent
vertebrate calpain with a novel C-terminal domain. Mamm. Genome. 10, 318–321.
Goll, D. E., Thompson, V. F., Li, H., Wei, W., and Cong, J. (2003). The calpain
system. Physiol. Rev. 83, 731–801.
Page | 61
Guroff, G., and Guroff, G. (1964). A neutral calcium-activated proteinase from the
soluble fraction of rat brain. J. Biol.Chem. 239, 149.
Horikawa, Y., Oda, N., Cox, N.J. et al. (2000). Genetic variation in the gene encoding
calpain-10 is associated with type 2 diabetes mellitus. Nat. Genet. 26, 163–175.
Hosfield, C. M., Moldoveanu, T., Davies, P. L., Elce, J. S., and Jia, Z. (2001). Calpain
mutants with increased Ca2+ sensitivity and implications for the role of the C(2)like domain. J. Biol.Chem. 276, 7404–7407.
Hubbard, S.R., and Miller, W.T. (2007). Receptor tyrosine kinases: mechanisms of
activation and signaling. Curr Opin Cell Biol. 19(2), 117-123.
Igarashi, Y., Eroshkin, A., Gramatikova, S., Gramatikoff, K., Zhang, Y., Smith, J.W.,
Osterman, A.L., and Godzik, A. (2007). CutDB: a proteolytic event database.
Nucleic Acids Research. 35, D546–D549.
Ishiura, S., Murofushi, H., Suzuki, K., and Imahori, K. (1978). Studies of a calciumactivated neutral protease from chicken skeletal muscle. I. Purification and
characterization. J. Biochem. 84, 225-230.
Joachims, T. (1999). Making large-Scale SVM Learning Practical. Advances in
Kernel Methods - Support Vector Learning, B. Schölkopf and C. Burges and A.
Smola (ed.).MIT-Press.
Kamei, M., Webb, G. C., Young, I. G., and Campbell, H. D. (1998). SOLH, a human
homologue of the Drosophila melanogaster small optic lobes gene is a member of
the calpain and zinc-finger gene families and maps to human chromosome 16p13.3
near CATM (cataract with microphthalmia). Genomics. 51, 197–206.
Liu, Z., Gao, X., Cao, J., Ma, Q., Ren, J., and Xue, Y. (2010). GPS-CCD: A novel
computational program for the prediction of calpain cleavage sites. Retrieved
December 30, 2010, from: http://ccd.biocuckoo.org/
Manning, G., Whyte, D.B., Martinez, R., Hunter, T., and Sudarsanam, S. (2002).The
protein kinase complement of the human genome. Science. 298, 1912–1934.
Molinari, M., Anagli, J., and Carafoli, E. (1995). PEST sequences do not influence
substrate susceptibility to calpain proteolysis. J Biol Chem. 270(5), 2032-2035.
Nakagawa, T., and Yuan, J. (2000). Cross-talk between two cysteine protease
families. Activation of caspase-12 by calpain in apoptosis. J Cell Biol. 150, 887–
894.
Noble, W. S. (2006). What is a support vector machine? Nature Biotechnology. 24,
1565-1567.
Ohno, S., Emori, Y., Imajoh, S., Kawasaki, H., Kisaragi, M., and Suzuki, K. (1984).
Evolutionary origin of a calcium-dependent protease by fusion of genes for a thiol
protease and a calcium binding protein? Nature. 312, 566-570.
Page | 62
Overall, C.M., Tam, E.M., Kappelhoff, R., Connor, A., Ewart, T., Morrison, C.J.,
Puente, X., López-Otín, C., and Seth, A. (2004). Protease degradomics: mass
spectrometry discovery of protease substrates and the CLIP-CHIP, a dedicated
DNA microarray of all human proteases and inhibitors. Biol Chem. 385(6), 493504.
R Development Core Team. (2010). R: A language and environment for statistical
computing. Retrieved June 1, 2010, R Foundation for Statistical Computing,
Vienna, Austria, from: http://www.R-project.org/
Ren, J., Gao, X., Jin, C., Zhu, M., Wang, X., Shaw, A., Wen, L., Yao, X., and Xue, Y.
(2009). Systematic study of protein sumoylation: Development of a site-specific
predictor of SUMOsp 2.0. Proteomics. 9(12), 3409-3412.
Ren, J., Wen, L., Gao, X., Jin, C., Xue, Y., and Yao, X. (2008). CSS-Palm 2.0: an
updated software for palmitoylation sites prediction. Protein End Des Sel. 21(11),
639-644.
Reverter, D., Sorimachi, H., and Bode, W. (2001). The structure of calcium free
human m-calpain. Implications for calcium activation and function. Trends
Cardiovasc Med. 11, 222–229.
Sakai, K., Akanuma, H., Imahori, K., and Kawashima, S. (1987). A unique specificity
of a calcium activated neutral protease indicated in histone hydrolysis. J Biochem.
101(4), 911-918.
Sasaki, T., Kikuchi, T., Yumoto, N., Yoshimura, N., and Murachi, T. (1984).
Comparative specificity and kinetic studies on porcine calpain I and calpain II with
naturally occurring peptides and synthetic fluorogenic substrates. J Biol Chem.
259(20), 12489-12494.
Schneider, T.D., and Stephens, R.M. (1990). Sequence logos: A new way to display
consensus sequences. Nucleic Acids Res. 18, 6097–6100.
Shao, J., Xu, D., Tsai, S.N., Wang, Y., and Ngai, S.M. (2009). Computational
Identification of Protein Methylation Sites through Bi-Profile Bayes Feature
Extraction. PLoS One. 4, e4920.
Stabach, P.R., Cianci, C.D., Glantz, S.B., Zhang, Z., and Morrow, J.S. (1997). Sitedirected mutagenesis of alpha II spectrin at codon 1175 modulates its mu-calpain
susceptibility. Biochemistry. 36(1), 57-65.
Strobl, S., Fernandez-Catalan, C., Braun, M. et al. (2000). The crystal structure of
calcium-free human m-calpain suggests an electrostatic switch mechanism for
activation by calcium. Proc. Natl Acad. Sci. USA. 97, 588–592.
Suzuki, K. (1991). Nomenclature of calcium dependent proteinase. Biomed. Biochim.
Acta. 50, 483-484.
Page | 63
Tompa, P., Buzder-Lantos, P., Tantos, A., Farkas, A., Szilagyi, A., Banoczi, Z.,
Hudecz, F., and Friedrich, P. (2004). On the sequential determinants of calpain
cleavage. J. Biol. Chem. 279, 20775–20785.
Uniprot Consortium. (2010). The Universal Protein Resource in 2010. Nucleic Acids
Res. 38, D142–D148.
Vapnik, V. (1995). The Nature of Statistical Learning Theory. New York: Springer
Verlag.
Vosler, P. S., Brennan, C. S., and Chen, J. (2008). Calpain-Mediated Signaling
Mechanisms in Neuronal Injury and Neurodegeneration. Mol Neurobiol. 38, 78–
100.
Wang, K.K., Villalobo, A., and Roufogalis, B.D. (1989). Calmodulin-binding proteins
as calpain substrates. Biochem J. 262(3), 693-706.
Wee, L.J., Tan, T.W., and Ranganathan, S. (2006). SVM-based prediction of caspase
substrate cleavage sites. BMC Bioinformatics.7 (Suppl 5):S14
Wee, L.J., Tong, J.C., Tan, T.W., Ranganathan, S. (2009). A multi-factor model for
caspase degradome prediction. BMC Genomics.10 (Suppl 3):S6.
Xue, Y., Ren, J., Gao, X., Jin, C., Wen, L., and Yao X. (2008). GPS 2.0, a tool to
predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics.
7(9), 1598-1608.
Zhang, S.W., Pan, Q., Zhang, H.C., Zhang, Y.L., and Wang, H.Y. (2003).
Classification of protein quaternary structure with support vector machine.
Bioinformatics. 19, 2390-2396.
Page | 64
APPENDIX A
Page | 65
Table A-1: Dataset of calpain substrate cleavage sites (for cross-validation and SVM training).
Uniprot ID
P1 Position1
Cleavage Site2
ACAN
P13608
1229
EDLS-VLPS
ACAN
P13608
1287
EDLS-VLPS
ACAN
P13608
1346
EDLS-VLPS
ACAN
P13608
474
APGA-AEVP
ACAN
P13608
719
PGVA-AVPI
ACAN
P13608
365
FGVG-GEED
ACAN
P13608
1307
EDLG-VLPS
ACAN
P16112
1411
EDLS-GLPS
ACAN
P16112
954
GDLS-GLPS
ACAN
P16112
1681
PDLS-GQPS
ACAN
P16112
1452
TDLS-GLPS
ACAN
P16112
1431
GDLS-GVPS
ACAN
P16112
973
GDLS-GLPS
P68133/P62736/P60709
39, 39, 37
IVGR-PRHQ
ACTN1
P12814
243
TYVS-SFYH
ACTN1
P12814
246
SSFY-HAFS
ACTN4
O43707
265
SSFY-HAFS
Agt
P01015
28
DRVY-IHPF
Aifm1
Q9JM53
103
GLGL-SPEE
Aifm1
Q9Z0X1
102
MGLG-LSPE
Aifm1
Q9Z0X1
118
SATE-GGSV
AMPD1
P23109
85
VNLS-IPLS
AMPD1
P23109
97
TKLS-HIDE
AMPH
P49418
377
SPMS-QTLP
AMPH
P49418
392
TDLV-QPAS
AMPH
P49418
454
DLGM-DTRA
AMPH
P49418
333
PEIS-VTTP
AMPH
P49418
478
AAVG-TLVS
AMPH
P49418
593
PIQD-PQPT
AMPH
P49418
531
EELE-ATVP
AMPH
P49418
527
QPEA-EELE
AMPH
P49418
609
ADQL-ASAR
Ankrd2
Q9WV06
77
EEKR-LGVQ
ANXA1
P04083
26
QTVK-SSKG
Calpain Substrate
ACTA1/ACTA2/ACTB
ATG5
Q9H1Y0
193
YQTT-TERP
Q01814-1/6
1124, 1079
RELR-RGQI
ATXN3
P54252
200
AQLK-EQRV
ATXN3
P54252
60
DYRT-FLQQ
BAX
Q07812
33
FIQD-RAGR
BAX
Q07812
28
LLLQ-GFIQ
BCL2
P10415
73
SPLQ-TPAA
BCL2L1
Q07817
42
EGTE-SEME
BID
P55957
54
WEGY-DELQ
ATP2B2
Page | 66
Uniprot ID
P1 Position1
Cleavage Site2
P55957
70
SRLG-RIEA
Camk4
P08414/Q16566
199, 203
TVCG-TPGY
Camk4
P08414
23
STEN-LVPD
CANP B
Q9VT65
74
HAQN-ASYA
CAPN1
P07384
27
RELG-LGRH
Capn3
P16259
296
NMDN-SLLR
Capn3
P16259
322
PVQY-ETRM
CASP14
P31944
152
VMVI-KDSP
CASP-7
P55210
36
PSLF-SKKK
CASP-7
P55210
45
NVTM-RSIK
CASP-7
P55210
47
TMRS-IKTT
CASP9
P55211-1/2
330
DQLD-AISS
CASP9
P55211-1
143
GALE-SLRG
CASP9
P55211-1
120
KPEV-LRPE
CASP9
P55211-1/2
115
RPEI-RKPE
CASP9
Calpain Substrate
BID
P55211-2
120
KPEV-LRPE
CDK5R1
Q15078
98
LSTF-AQPP
CDK5R2
Q13319
100
QQRN-RENL
CDK5R2
Q13319
108
LRKG-RDPP
CDK5R2
Q13319
105
ENLL-RKGR
CDKN2D
P55273
64
LKQG-ASPN
CDKN2D
P55273
29
RLLH-RELV
CDKN2D
P55273
113
PIHL-AVQE
CDKN2D
P55273
127
SFLA-AESD
CDKN2D
P55273
47
TALQ-VMMF
CDKN2D
P55273
25
QEVR-RLLH
CRYBA1
P11843
22
AQTN-PMPG
Ctnnb1
Q02248
28
WQQQ-SYLD
Ctnnb1
Q02248
29
QQQS-YLDS
Ctnnb1
Q02248
30
QQSY-LDSG
CTTN
Q14247-1
358
ENLA-KEKE
CTTN
Q14247-1
351
SNIR-ANFE
CTTN
Q14247-1
336
AYQK-TVPV
DMD
P11532
690
TVTT-REQI
DMD
P11532
1992
MPLE-ISYV
EGFR
P00533
1030
PSTS-RTPL
EGFR
P00533
1086
DDTF-LPVP
EGFR
P00533
1151
NSTF-DSPA
EGFR
P00533
683
RRLL-QERE
EGFR
P00533
733
LWIP-EGEK
EGFR
P00533
1185
KPNG-IFKG
F2R
P25116
32
PESK-ATNA
F2R
P25116
76
SINK-SSPL
F2RL1
P55085
58
VETV-FSVD
Page | 67
Uniprot ID
P1 Position1
Cleavage Site2
F2RL1
P55085
71
VLTG-KLTT
FADK 1
Q05397
745
YQVS-GYPG
FCGR2A
P12318
263
EPPG-RQMI
FCGR2A
P12318
268
QMIA-IRKR
FCGR2A
P12318
255
DPVK-AAQF
FLNA
P21333
1761
APQY-TYAQ
Fos
P12841
90
PSQT-RAPH
GAD2
Q05329
69
AAAR-KAAC
Gap43
P07936
40
KIQA-SFRG
Gcg
P06883
67
KYLD-SRRA
Gcg
P06883
69
LDSR-RAQD
Gcg
P06883
77
FVQW-LMNT
Gcg
P06883
74
AQDF-VQWL
GJA8
P55917
300
SPLS-AKPF
Gnrh1
P07490
28
HWSY-GLRP
Gnrh1
P07490
29
WSYG-LRPG
Grin2a
Q00959
1278
NALQ-FQKN
Grin2a
Q00959
1329
GSLF-SVPS
Grm1
P23385
936
LTKS-YQGS
GRM1
Q13255
936
LTKS-YQGS
HIST1H2BC
P62808
106
LPGE-LAKH
HIST1H2BC
P62808
42
SVYV-YKVL
HIST1H2BC
P62808
81
ASRL-AHYN
HIST1H2BC
P62808
64
GIMN-SFVN
HIST1H2BC
P62808
20
KAVT-KAQK
HIST1H2BC
P62808
101
AVRL-LLPG
HIST1H2BC
P62808
46
YKVL-KQVH
HIST1H2BC
P62808
40
SYSV-YVYK
HIST1H2BC
P62808
96
REIQ-TAVR
HIST1H2BC
P62808
105
LLPG-ELAK
HIST1H2BC
P62808
53
HPDT-GISS
HTT
P42858
534
SHSS-SQVS
HTT
P42858
467
SALT-ASVK
IGFBP-2
P18065
202
TEQH-RQMG
IGFBP-3
P17936
175
HPLH-SKII
IGFBP4
P22692
143
QKHF-AKIR
IGFBP4
P22692
107
AEIE-AIQE
IGFBP4
P22692
23
LGDE-AIHC
IGFBP4
P22692
159
MKVN-GAPR
IGFBP5
P24593
161
KKLT-QSKF
IGFBP5
P24593
172
AENT-AHPR
IGFBP5
P24593
22
QSLG-SFVH
IL1A
P01583
118
PFSF-LSNV
INS
P01317
29
VNQH-LCGS
Calpain Substrate
Page | 68
Uniprot ID
P1 Position1
Cleavage Site2
INS
P01317
40
EALY-LVCG
INS
P01317
37
HLVE-ALYL
ITGB1
P05556-5
777
KWDT-QENP
ITGB1
P05556-5
772
EKMN-AKWD
ITGB1
P05556
771
KEKM-NAKW
ITGB1
P05556-5
767
AKFE-KEKM
ITGB1
P05556-5
778
WDTQ-ENPI
ITGB1
P05556-5
771
KEKM-NAKW
ITGB1
P05556
778
WDTG-ENPI
ITGB3
P05106
767
KWDT-ANNP
ITGB3
P05106
768
WDTA-NNPL
ITGB3
P05106
761
EERA-RAKW
ITGB7
P26010
770
QLNW-KQDS
ITGB7
P26010
774
KQDS-NPLY
ITGB7
P26010
769
QQLN-WKQD
ITGB7
P26010
766
KEQQ-QLNW
ITGB7
P26010
760
EYSR-FEKE
ITGB7
P26010
765
EKEQ-QQLN
ITGB7
P26010
773
WKQD-SNPL
Jun
P17325
90
HITT-TPTP
Jun
P17325
62
DLLT-SPDV
Jun
P17325
164
ASLH-SEPP
Jun
P17325
42
TLNL-ADPV
KRT18
P05783
78
GGIQ-NEKE
KRT18
P05783
253
ADIR-AQYD
KRT18
P05783
186
HGLR-KVID
KRT18
P05783
80
IQNE-KETM
KRT18
P05783
236
LTVE-VDAP
KRT18
P05783
305
TELR-RTVQ
KRT18
P05783
64
GGLA-TGIA
KRT18
P05783
290
AEVG-AAET
KRT18
P05783
137
EDLR-AQIF
KRT18
P05783
59
GGMG-SGGL
KRT18
P05783
284
VVTT-QSAE
KRT8
P05787
444
SSFG-SGAG
KRT8
P05787
79
LVLE-VDPN
KRT8
P05787
77
SPLV-LEVD
KRT8
P05787
75
LLSP-LVLE
KRT8
P05787
440
YSLG-SSFG
KRT8
P05787
236
RELQ-SQIS
Marcks
P26645
127
SSTS-SPKA
MARP2/ANKRD2
Q9GZV1
103
LDLR-REII
MBP
P02687
93
KNIV-TPRT
MBP
P02687
96
VTPR-TPPP
Calpain Substrate
Page | 69
Uniprot ID
P1 Position1
Cleavage Site2
MBP
P02686-1/2/3/4/5
152
LATA-STMD
MBP
P02686-1
204
AHYG-SLPQ
MBP
P02686-3/4
50
GGDR-GAPK
MBP
P02686-3/4
97
AHYG-SLPQ
MBP
P02686-1/2
157, 24
TMDH-ARHG
MBP
P02686-1/2/3/4/5
161, 28
ARHG-FLPR
MBP
P02686-1/3/4/5/6
279, 172,161,146,135
KGVD-AQGT
MBP
P02686-1/3/4/5
213, 106,106,80,80
SHGR-TQDE
MBP
P02686-1/3/5
231, 124, 98
VTPR-TPPP
MBP
P02686-1/3/5
241, 134, 108
GKGR-GLSL
MBP
P02686-1/3/5
265, 158, 132
GGRA-SDYK
MBP
P02686-1/3/5
244, 137, 111
RGLS-LSRF
MBP
P02686-4/6
147, 121
GGRA-SDYK
MBP
P02686-4/6
124, 98
VTPR-TPPP
MIP
Q6RZ07
239
ILKG-TRPS
MIP
P30301
237
LSVL-KGAK
MIP
P30301
238
SVLK-GAKP
MIP
Q6RZ07
238
SILK-GTRP
MIP_RAT
P09011
236
SILK-GARP
Calpain Substrate
Mtap2
P15146-3
99
QVVT-AEAV
MYO5A
Q02440
1140
LPLR-MEEP
MYOC
Q99972
226
PASR-ILKE
NEFM
O77788
467
EDEK-SEME
NF2
P35240-1/2
298
LILQ-LCIG
NF2
P35240-1/2/6
294, 294, 252
RVNK-LILQ
NFKBIA
P25963
50
KELQ-EIRL
PARP1
P18493
502
GKSG-AAPS
PARP1
P18493
384
AAVH-SGPP
PARP1
P18493
658
KKLT-VNPG
PDE1A
P54750
126
HAVQ-AGIF
PDE1A
P14100
126
HVVQ-AGIF
Pdyn
NA
207
GFLR-RIRP
Pdyn
NA
214
PKLK-WDNQ
PHKG
P00518
303
SPRG-KFKV
Plasmepsin-1
P39898
123
PHLG-NAGD
Plasmepsin-2
P46925
124
NYLG-SSND
PLCB1
P10894
880
QALH-SQPA
Ppp3ca
P63329
392
AAAR-KEVI
Ppp3ca
P63329
424
LTLK-GLTP
PPP3CA
Q08209
501
SINK-ALTS
Prkca
P05696
309
EKAK-LGPA
Prkca
P05696
316
AGNK-VISP
Prkca
P05696
324
SEDR-KQPS
Prkcb
P68403
311
AKIG-QGTK
Page | 70
Uniprot ID
P1 Position1
Cleavage Site2
Prkcg
P63319
338
KRCF-FGAS
Prkcg
P63319
321
GPSS-SPIP
PTBP1
P26599
165
LALA-ASAA
PTBP1
P26599
163
GNLA-LAAS
PTPRN
Q16849
659
SVSS-QFSD
PTRF
Q6NZI2
370
PDVH-ALLE
RB1
P06400
810
SPLK-SPYK
RCAN1
P53805-2
133
DLLY-AISK
RGP51
Q6QUW1
58
QQLS-SSGI
RYR1
P11716
1400
AMMT-QPPA
RYR1
P11716
2843
RKIS-QTAQ
SAG
P08168
377
NFVF-EEFA
SAG
P08168
380
FEEF-ARQN
SLC6A3
Q01959
71
DFLL-SVIG
SLC6A3
Q01959
43
VQLT-SSTL
Slc6a5
P58295
164
VVLG-TDGI
Slc6a5
P58295
156
WVNM-SQST
Slc6a9
Q63322/P28572-1
26
QNLT-RGNW
Slc6a9
Q63322/P28572-1
26
QNLT-RGNW
Slc6a9
Q63322/P28572-2
31
QNLT-RGNW
Slc6a9
Q63323/P28572-1
31
QNLT-RGNW
SLC8A3
P57103
512
PRAV-LASP
SLC8A3
P57103
510
PLPR-AVLA
SLC8A3
P57103
370
NILK-KHAA
SLC8A3
P57103
504
AIFN-SLPL
SMN1
Q16637
193
WNSF-LPPP
SMN1
Q16637
192
PWNS-FLPP
SNCA
P37840
57
TVAE-KTKE
SNCA
P37840
83
KTVE-GAGS
SNCA
P37840
75
TGVT-AVAQ
SNCA
P37840
74
VTGV-TAVA
SNCA
P37840
114
GILE-DMPV
SPTAN1
Q13813
1230
QLLG-SAHE
SPTAN1
Q13813
1176
QEVY-GMMP
SPTB
P11277
2058
EKST-ASWA
SPTB
P11277
2061
TASW-AERF
SPTBN1
Q01082
1440
EELQ-SQAQ
SPTBN1
Q01082
1467
QTKF-MELL
SPTBN1
Q01082
1482
HNLL-ASKE
Calpain Substrate
SPTBN1
Q01082
1447
QALS-QEGK
SPTBN1
Q01082-1/3
2066, 2053
EKSA-ATWD
TH
P17289
30
EAIM-SPRF
TH
P17289
22
SELD-AKQA
Tln1
P26039
433
TVLQ-QQYN
Page | 71
Uniprot ID
P1 Position1
Cleavage Site2
Tnnt2
P50752
81
KPSR-LFMP
TNNT3
P02641
65
PKLT-APKI
TOP1
P11387
183
DKDK-KVPE
TOP1
P11387
158
ADYK-PKKI
TP53
P04637
20
ETFS-DLWK
TPM1
P58772
256
IDDL-EDEL
TPM1
P58772
223
KYEE-EIKV
TPM1
P58772
183
EERA-ELSE
TPM1
P58772
241
RAEF-AERS
TPM1
P58772
205
NNLK-SLEA
TPM1
P58772
27
QAEA-DKKA
TPM1
P58772
204
TNNL-KSLE
tra-2
P34709
1088
ATKQ-MFES
TTN
Q9Y6L9,Q8WZ42-4
8651
QRLS-QTEP
TTN
Q9Y6L9,Q8WZ42-4
8506
IHQK-GDEA
TTN
Q9Y6L9,Q8WZ42-4
8563
MLKK-TPIL
TTN
Q9Y6L9,Q8WZ42-4
8652
RLSQ-TEPV
Ttn
A2ASS6
9828
IPDS-RVPI
Vim
P20152
53
RSLY-SSSP
Vim
P20152
92
ADAI-NTEF
Vim
P20152
71
VRLR-SSVP
Vim
P20152
33
YVTT-STRT
Vim
P20152
64
YVTR-SSAV
Vim
P20152
38
TRTY-SLGS
Vim
P20152
41
YSLG-SALR
Vim
P20152
266
PDLT-AALR
VWF
P04275
1913
TLLK-SHRV
VWF
P04275
763
RSKR-SLSC
Calpain Substrate
Cleavage sites are reported as octapeptides in the order: P4-P3-P2-P1-P1’-P2’-P3’-P4’.
Cleavage sites containing exact sequence information but originating from multiple
isoforms (if any) are demarcated by commas.
1
2
Position of the P1 amino acid in the protein sequence as reported in Uniprot.
Page | 72
Table A-2: Dataset of calpain substrate cleavage sites (for independent out-of-sample testing).
Calpain Substrate
Uniprot ID
P1 Position1
Cleavage Site2
30K, Calpain regulatory subunit
O42134
87
GFGL-DTCR
ACAN
P13608
1249
EDLS-VLPS
ACAN
P16112
365
FGVG-GEED
ACAN
P16112
1353
GDLS-GLPS
ACAN
P16112
1472
EDLS-GLPS
ACAN
P16112
709
PGVA-AVPV
Ap2b1
P62944
677
PATF-APSP
AP2B1
Q3ZB97
691
PATF-APSP
Q01814-1/6
1135, 1090
RGLN-RIQT
ATXN3
P54252
260
MQGS-SRNI
Bcl2l1
Q64373
60
WHLA-DSPA
CANP B
Q9VT65
224
PENQ-NMFW
Capn3
P16259
591
ISVD-RPVK
Capn3
P16259
274
NMTY-GTSP
COPB1
P53618
528
SALS-SSRP
Ctnnb1
Q02248
95
QRVR-AAMF
Cttn
Q60598
358
ENLA-KERE
CTTN
Q14247-1
346
VTSK-TSNI
EGFR
P00533
1059
QSCP-IKED
EZR
P15311
467
HLVM-TAPP
F2RL1
P55085
59
ETVF-SVDE
F2RL1
P55085
45
VDGT-SHVT
FLNC
Q14315
2626
SSYS-SIPK
Gcg
P06883
79
QWLM-NTKR
Gfap
P03995
56
GALN-AGFK
Gfap
P03995
29
RQLG-TMPR
GJA8
P55917
290
PLTE-VGMV
INS
P01317
32
HLCG-SHLV
INS
P01317
50
GFFY-TPKA
ITGB1
P05556
777
KWDT-GENP
ITGB2
P05107
744
EKLK-SQWN
ITGB7
P26010
778
NPLY-KSAI
KRT18
P05783
286
TTQS-AEVG
KRT18
P05783
30
RPVS-SAAS
KRT18
P05783
285
VTTQ-SAEV
KRT8
P05787
73
QSLL-SPLV
KRT8
P05787
72
NQSL-LSPL
LCP1
P13796
109
TSEQ-SSVG
ATP2B2
MBP
P02687
68
THYG-SLPQ
MBP
P02686-1/5/6
183, 50
GGDR-GAPK
MBP S
P02688
114
VHFF-KNIV
NEFM
O77788
516
SPVK-ATAP
Page | 73
Uniprot ID
P1 Position1
Cleavage Site2
PARP1
P18493
480
THLL-SPWG
Prkcb
P68403
320
PEEK-TANT
PTPRN
Q16849
608
RQQD-KERL
PTRF
Q6NZI2
30
AGAQ-AAEE
SNCA
P37840
73
VVTG-VTAV
TP53
P04637
25
LWKL-LPEN
TPM1
P58772
208
KSLE-AQAE
Vim
P20152
21
SGTS-SRPS
Calpain Substrate
Cleavage sites are reported as octapeptides in the order: P4-P3-P2-P1-P1’-P2’-P3’-P4’.
Cleavage sites containing exact sequence information but originating from multiple
isoforms (if any) are demarcated by commas.
1
2
Position of the P1 amino acid in the protein sequence as reported in Uniprot
Page | 74
APPENDIX B
Page | 75
B-1: Grid search optimization tables obtained for simple binary encoded symmetrical
subsequence windows (P4P4’, P8P8’, P12P12’, P16P16’ and P20P20’) and asymmetrical
subsequence windows (P4P12’ and P12P4’).
Optimal γ and C values are highlighted in blue.
C
γ
0.001
0.01
0.1
1
10
100
0.001
66.15
65.98
61.34
51.89
51.89
51.72
0.01
66.15
65.98
61.34
51.89
51.89
51.72
0.1
66.15
65.98
61.34
51.89
51.89
52.75
1
66.15
68.04
69.93
55.15
53.44
53.44
10
68.73
68.56
69.42
55.33
53.44
53.44
100
Binary P4P4’
69.07
68.21
69.42
55.33
53.44
53.44
0.001
0.01
0.1
1
10
100
0.001
67.18
65.98
53.44
52.06
51.20
51.20
0.01
67.18
65.98
53.44
52.06
51.20
51.20
0.1
67.18
65.98
53.44
52.06
52.06
52.06
1
67.18
70.27
72.16
53.95
52.92
52.92
10
71.48
69.07
72.68
53.95
52.92
52.92
100
Binary P8P8’
68.90
63.75
72.68
53.95
52.92
52.92
0.001
0.01
0.1
1
10
100
0.001
63.40
61.86
52.23
50.86
50.69
50.69
0.01
63.40
61.86
52.23
51.37
50.69
50.69
0.1
63.40
61.86
52.23
51.37
51.37
51.37
1
63.40
71.99
70.62
52.92
52.41
52.41
10
71.48
68.38
72.34
53.44
52.41
52.41
100
68.04
Binary P12P12’
66.15
72.34
53.44
52.41
52.41
0.001
0.01
0.1
1
10
100
0.001
57.39
55.67
51.20
50.69
50.69
50.69
0.01
57.39
55.67
51.20
50.69
50.69
50.69
0.1
57.39
55.67
51.20
50.86
50.86
50.86
1
57.39
70.62
61.51
52.23
51.03
51.03
10
69.93
66.49
65.46
52.58
51.03
51.03
100
65.46
Binary P16P16’
66.49
65.46
52.58
51.03
51.03
C
γ
C
γ
C
γ
Page | 76
C
γ
0.001
0.01
0.1
1
10
100
0.001
55.67
54.12
51.55
51.03
51.03
52.23
0.01
55.67
54.12
51.55
51.03
51.03
52.23
0.1
55.67
54.12
51.55
51.03
51.03
52.23
1
55.67
69.24
51.19
52.23
51.03
52.23
10
67.18
65.81
56.87
52.23
51.03
52.23
100
62.89
Binary P20P20’
65.81
56.87
52.23
51.03
52.23
0.001
0.01
0.1
1
10
100
0.001
66.67
67.01
54.30
51.37
50.69
50.69
0.01
66.67
67.01
54.30
51.37
50.69
50.69
0.1
66.67
67.01
54.30
51.37
51.37
51.37
1
66.67
69.07
69.59
53.44
52.41
52.41
10
68.56
68.90
69.24
53.61
52.41
52.41
69.42
64.26
69.24
53.61
52.41
52.41
0.001
0.01
0.1
1
10
100
0.001
62.71
60.31
53.44
51.72
51.72
51.72
0.01
62.71
60.31
53.44
51.72
51.72
51.72
0.1
62.71
60.31
53.44
51.72
52.58
52.58
1
62.71
71.13
70.79
54.12
53.09
53.09
10
71.31
68.04
71.82
54.12
53.09
53.09
67.53
64.43
71.82
54.12
53.09
53.09
C
γ
100
Binary P4P12’
C
γ
100
Binary P12P4’
Page | 77
B-2: Grid search optimization tables obtained for Bayes Feature Extraction (BFE)
encoded symmetrical subsequence windows (P4P4’, P8P8’, P12P12’, P16P16’ and P20P20’)
and asymmetrical subsequence windows (P4P12’ and P12P4’).
Optimal γ and C values are highlighted in blue.
C
γ
0.001
0.01
0.1
1
10
100
0.001
63.92
69.93
52.58
51.89
51.89
51.72
0.01
63.92
69.93
52.58
51.89
51.89
51.72
0.1
69.07
71.13
52.58
51.89
52.75
52.75
1
74.57
71.99
55.50
53.61
53.44
53.44
10
73.88
67.87
57.04
53.61
53.44
53.44
100
69.59
67.70
57.04
53.61
53.44
53.44
0.001
0.01
0.1
1
10
100
0.001
66.49
70.96
52.58
51.20
51.20
51.20
0.01
66.49
70.96
52.58
51.20
51.20
51.20
0.1
70.10
70.96
52.58
52.06
52.06
52.06
1
76.63
73.02
54.30
52.92
52.92
52.92
10
76.46
72.34
54.30
52.92
52.92
52.92
100
71.48
72.34
54.30
52.92
52.92
52.92
0.001
0.01
0.1
1
10
100
0.001
67.01
53.78
51.72
50.69
50.69
50.69
0.01
67.01
53.78
51.72
50.69
50.69
50.69
0.1
70.27
53.78
51.72
51.37
51.37
51.37
1
79.21
71.82
53.26
52.41
52.41
52.41
10
78.87
72.16
53.44
52.41
52.41
52.41
100
75.26
72.16
53.44
52.41
52.41
52.41
0.001
0.01
0.1
1
10
100
0.001
66.84
51.20
50.69
50.69
50.69
50.69
0.01
66.84
51.20
50.69
50.69
50.69
50.69
0.1
70.45
51.20
50.86
50.86
50.86
50.86
1
80.58
62.71
51.89
51.03
51.03
51.03
10
78.01
67.70
51.89
51.03
51.03
51.03
100
78.01
67.70
51.89
51.03
51.03
51.03
Bayes P4P4’
C
γ
Bayes P8P8’
C
γ
Bayes P12P12’
C
γ
Bayes P16P16’
Page | 78
C
γ
0.001
0.01
0.1
1
10
100
0.001
68.04
51.37
50.17
49.66
52.34
49.31
0.01
68.04
51.37
50.17
49.66
52.34
49.31
0.1
70.27
51.37
50.17
49.66
52.34
49.31
1
79.55
56.53
51.37
49.66
52.34
49.31
10
78.52
57.73
52.23
50.17
52.34
49.31
100
78.01
57.73
52.23
50.17
52.34
49.31
0.001
0.01
0.1
1
10
100
0.001
64.78
70.45
51.89
50.69
50.69
50.69
0.01
64.78
70.45
51.89
50.69
50.69
50.69
0.1
69.42
70.45
51.89
51.37
51.37
51.37
1
78.18
73.02
53.44
52.41
52.41
52.41
10
78.18
71.99
53.61
52.41
52.41
52.41
100
74.91
71.99
53.61
52.41
52.41
52.41
0.001
0.01
0.1
1
10
100
0.001
66.49
70.27
51.72
51.72
51.72
51.72
0.01
66.49
70.27
51.72
51.72
51.72
51.72
0.1
70.45
70.27
51.72
52.58
52.58
52.58
1
76.29
72.68
54.81
53.09
53.09
53.09
10
76.80
73.02
54.98
53.09
53.09
53.09
100
72.34
73.02
54.98
53.09
53.09
53.09
Bayes P20P20’
C
γ
Bayes P4P12’
C
γ
Bayes P12P4’
Page | 79
APPENDIX C
Page | 80
Figure C-1: Combined ROC curves and AROC scores generated for simple binary
encoded symmetrical and asymmetrical subsequence windows.
Page | 81
Figure C-2: Combined ROC curves and AROC scores generated for BFE encoded
symmetrical and asymmetrical subsequence windows.
Page | 82
APPENDIX D
Page | 83
For Tables D1-3, maximum intensity scores for each residue position are highlighted in yellow.
Table D-1: Amino acid intensities generated for the 40-mer positive dataset.
Page | 84
Table D-2: Amino acid intensities generated for the 40-mer negative dataset.
Table D-3: Calculated amino acid propensity, P .
X
Page | 85
Table D-4: Average P of each amino acid is calculated by averaging the P values of
X
X
the particular amino acid across all residue positions on the 40-mer peptides from the
experimentally-verified calpain cleavage sites (positive dataset), randomly generated
calpain non-cleavage sites (negative dataset) and calculated propensity P
X
.
Page | 86
APPENDIX E
Page | 87
Figure E1: Gantt chart of the BME499 Capstone project plan. Emphasis has been placed on critical stages of calpain substrate dataset collection,
implementation and testing of the SVM methodology and report writing.
Page | 88