Genotyping procedures - European Respiratory Journal
advertisement

Genotyping procedures
Extraction of genomic DNA from EDTA-anticoagulated whole blood was performed
manually using the PuregeneTM DNA Isolation Kit (Gentra Systems, Plymouth, MN,
USA) as described in detail elsewhere [1]. Genotyping of GSTT1, GSTM1 gene
deletions and GSTP1 p.Ile105Val SNP in the SAPALDIA cohort have been
previously reported [2].
Three single nucleotide polymorphisms (SNPs) of the HMOX1 gene were genotyped
using 5' nuclease real time PCR (TaqMan®) assay and fluorescently labeled allelespecific probes on the ABI Prism 7900 sequence detection system (Applied
Biosystems, Rotkreuz, Switzerland). The following primers and probes were used for
HMOX1 (rs2071746): 5’- AGCAA[T/A]AGCCTGGTGGG-3’,
forward 5’- CTGGGGTTGCTAAGTTCCTG -3’,
reverse 5’- CTGGGTGTGATTTTGCTCCT -3’
HMOX1 (rs5995098): 5’- TGCTTCATTAG[G/C]GTGAC -3’,
forward 5’- TGTGGCTTTCTCACTTGTGC -3’,
reverse 5’- CAGCCAGGGTACCACACTTT -3’
HMOX1 (rs735266): 5’- TCAGCGAAAAC[T/A]GATCT -3’,
forward 5’- GAGTAATCCTTTCCCGAGCC -3’,
reverse 5’- CGTGACTCAGCGAAAACAGA -3’.
The genotype call rate was >99%. Hardy–Weinberg equilibrium (HWE) using
Arlequin Version 2.000 was tested for all three SNPs which did not deviate from the
HWE [3]. A random sample of 10% of all DNA samples was re-genotyped showing a
reproducibility of >99.5%. Linkage disequilibrium between the SNPs was: D’=0.98
for rs2071746 and rs5995098, D’=0.97 for rs2071746 and rs735266 and D’=0.97 for
rs5995098 and rs735266. HMOX1 haplotypes were inferred using PHASE software
v2.1 based on the three SNPs in the following order: rs2071746, rs735266, rs5995098
[4]. Haplotypes with prevalences below 5% in our study population were not
analyzed.
The (GT)n promoter repeat in the HMOX1 gene was genotyped using an ABI Prism
3100 Genetic Analyzer (Applied Biosystems, Rotkreuz, Switzerland). Repeat
fragments were amplified using following primers:
forward 5’- CTCTGGAAGGAGCAAAAT C-3’,
reverse (6-FAM)-modified 5’- ACAAAGTCTGGCCATAGGAC-3’.
Fragment size analysis and allele attribution were performed using GeneMapper®
software v3.5 (Applied Biosystems, Rotkreuz, Switzerland) and subsequently each
genotype was visually verified. To confirm repeat allele attribution, a set of DNA
samples featuring homozygote genotypes covering the fragment range observed in our
population were sequenced. For comparability with the two published, population
based studies [5, 6], we categorised the HMOX1 promoter repeat alleles into three
subclasses according to the number of (GT)n repeats: short alleles, S (<=26 GT
repeats), medium alleles, M (27-32 GT repeats) and long alleles, L (>=33 GT
repeats). Subjects were then classified as having at least one long allele or not.
-1-
Glossary of specific genetic terms
Call rate
The call rate denominates the percentage of individual samples that have been
successfully genotyped on a specific polymorphism. It is thus a measure of the quality
of genotyping.
Hardy-Weinberg equilibrium (HWE)
The HWE describes how the three genotypes resulting from a single nucleotide
polymorphism (e.g. homozygous wild-type ‘aa’, heterozygous mutant ‘ab’, and
homozygous mutant ‘bb’) would be distributed within a population under the laws of
Mendelian inheritance.
If the frequency of allele ‘a’ in the population was denoted ‘p’ and the frequency of
allele ‘b’ ‘q’, then we would expect the following distribution of genotypes under
Hardy Weinberg equilibrium (HWE):
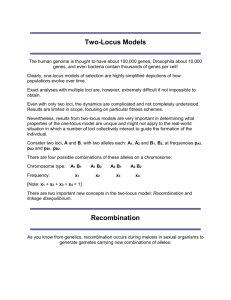
Genotype:
‘aa’
‘ab’
‘bb’
Expected frequency
p2
2*p*q
q2
Deviations from HWE in the distribution of genotypes can be observed because of
genotyping errors. The HWE is thus a measure of genotyping quality.
Alternatively, deviations from HWE can also be caused by ‘population stratification’.
Population stratification means admixture of a subgroup with different ethnic origin to
the study population. This could introduce confounding by ethnic background in
genetic analyses, if the ethnic subgroup is not equally distributed between the
outcome groups (the differences in genetic constitution observed between the
outcome groups are due to different ethnic background distributions and not to a
relationship of these genetic variants with the outcome).
Linkage disequilibrium (LD)
The linkage disequilibrium measures whether the combinations of alleles on two gene
loci located on the same chromosome are more frequently found together than would
be expected by chance. The chance that the alleles on two loci are passed on together
from one generation to the next on the same chromosome strand is a function of the
distance between the two loci and the rate of recombinations in the respective DNA
region during meiosis. If the alleles are more frequently found together than expected,
we speak of ‘linkage disequilibrium’ and the two loci are either likely to be relatively
close to each other or the recombination rate in the region is low. There are different
measures of linkage disequilibrium. A frequently used one is the measure D’
(pronounced ‘D prime’).
Let’s suppose we are interest in investigating haplotypes from two loci L1 and L2
which have the alleles L1: A, a and L2: B, b. We can now calculate whether the
observed frequency of the haplotypes differs from what we would expect based on the
frequency of the alleles by calculation of the disequilibrium measure D:
D = PAB – PA*PB, where P denotes the frequency of the respective allele.
Equivalent calculations are possible for the other haplotypes.
Linkage disequilibrium is present if D≠0.
But the maximum range of values D can take is from -0.25 to +0.25 (e.g. if only two
haplotypes were present and the underlying allele-frequencies are all 0.5). This range
is highly dependent on the frequency of the alleles (frequencies close to 0 or 1 are
-2-
particularly problematic). To solve this problem Lewontin [7] proposed the measure
D’, which is the D measure standardized by it’s maximum attainable value Dmax.
min{ PA*Pb, Pa*PB} if D>0
D’ = D / Dmax, where Dmax = or
min {PA*PB, Pa*Pb} if D<0
D’ then ranges from 0 to 1, 0 indicating no linkage disequilibrium and values close to
1 high linkage disequilibrium.
-3-
References
1.
Ackermann-Liebrich U, Kuna-Dibbert B, Probst-Hensch NM, Schindler C,
Felber Dietrich D, Stutz EZ, Bayer-Oglesby L, Baum F, Brandli O, Brutsche
M et al: Follow-up of the Swiss Cohort Study on Air Pollution and Lung
Diseases in Adults (SAPALDIA 2) 1991-2003: methods and
characterization of participants. Soz Praventivmed 2005, 50(4):245-263.
2.
Imboden M, Downs SH, Senn O, Matyas G, Brändli O, Russi EW, Schindler
C, Ackermann-Liebrich U, Berger W, Probst-Hensch NM: Glutathione Stransferase genotypes modify lung function decline in the general
population: SAPALDIA cohort study. Respiratory Research 2007, 8:2-2.
3.
Schneider S, Roessli D, Excoffier L: Arlequin ver 2000: a software for
population genetics data analysis. . In.: Genetics & Biometry Laboratory,
University of Geneva, Switzerland; 2000.
4.
Stephens M, Smith NJ, Donnelly P: A new statistical method for haplotype
reconstruction from population data. American journal of human genetics
2001, 68(4):978-989.
5.
Siedlinski M, van Diemen CC, Postma DS, Boezen HM: Heme oxygenase 1
variations and lung function decline in smokers: proof of replication.
Journal of Medical Genetics 2008, 45(6):400-400.
6.
Guénégou A, Leynaert B, Bénessiano J, Pin I, Demoly P, Neukirch F,
Boczkowski J, Aubier M: Association of lung function decline with the
heme oxygenase-1 gene promoter microsatellite polymorphism in a
general population sample. Results from the European Community
Respiratory Health Survey (ECRHS), France. Journal of medical genetics
2006, 43(8):e43-e43.
7.
Lewontin RC: On measures of gametic disequilibrium. Genetics 1988,
120(3):849-852.
-4-