Sample_Preparation_for_Microarray_Handbook
advertisement

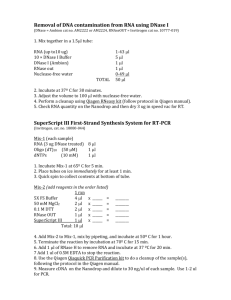
Functional Genomics Core Diabetes and Endocrinology Research Center Center for Molecular Studies in Digestive and Liver Disease Microarray Sample Preparation Handbook Director: Klaus Kaestner, PhD Marie Scearce, PhD John Brestelli Saki Arsenlis Location: 560 CRB Phone: 215-746-6372 Fax:215-573-5892 Email: mscearce@mail.med.upenn.edu 116100075 2/12/16 Kaestner Lab Table of Contents BASIC EXPERIMENTAL REQUIREMENTS: ....................................................................................... 3 ISOLATION OF TOTAL RNA FROM TISSUE BY ACID/PHENOL EXTRACTION ....................... 4 REAGENTS: ................................................................................................................................................ 4 PROTOCOL:................................................................................................................................................ 4 DNASE TREATMENT: ............................................................................................................................... 5 ISOLATION OF TOTAL RNA FROM TISSUE USING TRIZOL AND QIAGEN RNEASY MINICOLUMNS .................................................................................................................................................... 5 RNA ANALYSIS .......................................................................................................................................... 6 MEASURING THE CONCENTRATION OF RNA: ............................................................................................ 6 DENATURING AGAROSE GEL: ................................................................................................................... 6 Reagents: ........................................................................................................................................... 6 Protocol: ............................................................................................................................................. 7 2 116100075 2/12/16 Kaestner Lab Services: Comprehensive hybridization services…….$100/slide Includes basic data normalization and analysis PancChip version 2.2 …………………………$30/slide Custom arrays …………………………………Arranged upon request Basic Experimental Requirements: 4 replicate samples for each experimental condition tested 20ug of RNA (1-2ug/ul) per sample minimum (unless we are doing amplification) RNA must be DNAse treated Copies of the denaturing agarose gel to check RNA integrity OD 260 and OD 280 for each sample 3 116100075 2/12/16 Kaestner Lab Isolation of total RNA from Tissue By Acid/Phenol Extraction Reagents: Denaturing solution 4M Guanidium thiocyanate, 0.1 M Tris /Cl pH 7.5 (can keep this solution at room temperature) Prior to use add -mercaptoethanol to a final concentration of 1% Acid NaOAc solution 2M Sodium Acetate in DEPC-H20, pH = 4 in acetic acid Protocol: 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. 18. 19. 20. For approximately 1 g of tissue (approximately a cubic cm), arrange 50 ml Falcon tubes with 10 ml of denaturing solution in each on ice. Dissect out tissue and place immediately into cold denaturing solution. And immediately homogenize place homogenate on ice. Finish homogenizing rest of samples before proceeding on. Add 1 ml acidic NaOAc solution. Shake to mix. Add 10 mL phenol (pH4) (1 ml per ml lysate). Shake and invert to mix well. Add 4 ml chloroform:IAA [24:1] (0.4 ml per ml of lysate). Shake vigorously for 10 sec. Incubate for 15-30 minutes on ice. Centrifuge in falcon tubes at 6000 RPM for 20 minutes. Remove the aqueous phase to a new microcentrifuge tube. Add one volume isopropanol. Mix well by multiple inversion. Incubate at –20C for 2 or more hours (overnight is fine). Centrifuge at 6000 RPM for 30 minutes using Sorvall SLA-600TC. Aspirate the supernatant carefully. The pellet should be visible at this point. Suspend the pellet in 0.5 ml denaturing solution, and transfer to 2 microcentrifuge tubes. Add 50 L of acidic NaOAc to each. Mix well. Add 550 ml of 1:1 phenol:chloroform. to each Vortex and incubate 15 minutes on ice. Centrifuge at 13,000 rpm and 4C for 10 minutes. Remove the aqueous layer to a new microcentrifuge tube. Add 550 L of chloroform:IAA to samples. Repeat steps 16- 18. 4 116100075 2/12/16 21. 22. 23. 24. 25. 26. Kaestner Lab Add 550 L of isopropanol and mix well. Incubate at –80C for 10 minutes (or at -20C for > 1 hour). Centrifuge at 13,000 rpm and 4C for 10 minutes. Aspirate the supernatant, and wash the pellet with 0.5 ml 70% ethanol. Spin 3 minute Aspirate carefully. Pulse in centrifuge to pull any remaining fluid to bottom of the tube and aspirate and allow the pellet to dry for 10 minutes under the hood. Resuspend the pellet in 500 L of DEPC-treated H2O (1 ml for liver). If the RNA is difficult to resuspend, incubate RNA at 60C for 1 hr then vortex DNase Treatment: Reference: Wilson and Melton, Current Biology 1994, 4: 676-686. 1. 2. 3. Resuspend RNA pellet (approximately 70-140 ug) in 70 ul of DEPC-treated double distilled H2O 10 l 10 x DNase buffer (400 mM Tris/Cl pH 7.9, 100 mM NaCl, 60 mM MgCl2, 1 mM CaCl2) 10 l 20 mM DTT 2 l RNAsin, Promega 40U/l 8 l (80 units) RNase free DNase (BM) Incubate 30 minute at 37 oC Extract with phenol/chloroform, and precipitate with 0.1 ml 7.5 M ammonium acetate, 0.5 ml ethanol. Spin, wash with 70% ethanol, dry. Isolation of total RNA from tissue using TRIZOL 1. 2. 3. 4. 5. 6. 7. 8. Homogenize tissue(at least 15sec) samples in 1 ml TRIzol Reagent Add 20ug of Glycogen (may add up to 200ug for small concentration of tissue) directly to the TRIzol Add 200ul of chloroform per 1 ml of TRIzol Reagent. Shake tubes vigorously by hand for 15 sec and incubate at RT for 3 min Centrifuge for 30 min at 4 degrees Celsius 13K rpm. Transfer to layer and add 0.5ml of isopropanol per 1ml of Trizol Reagent used for the initial homogenization. Incubate on ice for 10 min and spin for 25 min at 4oC Wash the pellet with 1 ml of 75%ETOH, 25%DEPC water. Cf 25 min at 4oC. Re-suspend pellet in 300 ul of TES (10mM Tris pH7.5, 1mM EDTA pH8, 0.1-0.2% SDS). 5 116100075 2/12/16 9. 10. 11. 12. 13. 14. 15. Kaestner Lab Extract RNA with 600ul of phenol/Chloroform/isoamyl alcohol (25:24:1). Vortex vigorously and spin 5 min at 4oC. Carefully take the top phase, leaving any material at the interface. If there is a large interface, repeat extraction step. Add 1/10 volume, 3M Soduim Acetate pH7, and 3 volumes EtOH Percipitate at -80oC until use (at least 2 hrs, preferably overnight). Cf 30 min at 4oC Wash the pellet with 1 ml of 75%ETOH, 25%DEPC water. Cf 25 min at 4oC. resuspend to be >1ug/ul. RNA Analysis Measuring the concentration of RNA: 1. 2. 3. 4. 5. 6. Make a dilution of the RNA sample (usually 1:100) into TE buffer. Use spectrophotometer to read OD 260 If the OD 260 if higher than 1.0 or lower than 0.1 make a new dilution to bring the sample into a range where the accuracy of the readings is greatest and retake the OD 260 Measure the OD 280 and the OD260/280 ratio. The OD 280 measures contaminates in the sample. Therefore the OD260/280 ratio is an indication of RNA quality. The OD 260.OD280 range for high quality RNA is 1.8-2.0. Readings higher and lower indicate the presence of contaminates in the RNA and significantly reduces the accuracy of array analysis. Scan the purified RNA prep in a spectrophotometer (in TE buffer) to detect any phenol or protein contamination peaks. (Look for phenol peak at 270 nm) Do not continue with cDNA synthesis if ratio is less than 1.6 Denaturing Agarose Gel: Reagents: 1% Agarose Gel (1X TBE) TE Buffer (10 mM Tris-HCl; 1 mM EDTA pH8) Denaturing Sample Loading Buffer 2X TBE (pH8.3) 13%Ficoll (w/v) 0.01% Bromophenol Blue 7M Urea 6 116100075 2/12/16 Kaestner Lab Protocol: 1. 2. 3. 4. 5. Dilute 1-2 ug of RNA to a volume of 10 ul in DEPC water. Add equal volume Denaturing Loading Buffer to RNA Incubate RNA at 70 degrees Celsius for 10 minute; ice 5 minute Add entire sample to Gel; Run 80 V for 1 Hour. Integrity of RNA is assessed by visualizing 28S and 18 S Ribosomal bands. Smeared bands indicate degradation; High molecular weight bands indicate DNA contamination. 7