Supplementary Figure Legends - Word file
advertisement

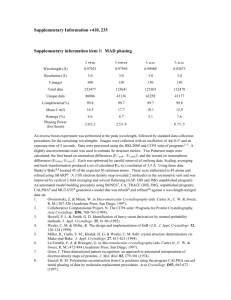
Toyoshima and Mizutani (2004-03-16597B) Supplementary Figure Legends 1 Supplementary Figure Legends Supplementary Figure 1. Configuration of cytoplasmic domains (A, N and P) in E2(TG) and E1·AMPPCP. Two structures are fitted with the top 12 residues of the M5 helix so that the P-domain has an equivalent orientation in either form. Several key residues and digestion sites by trypsin (T2) and proteinase K (PrtK) are marked. E486 appears to form a hydrogen bond with H190 in E2(TG) and with T171 in E1·AMPPCP. Supplementary Figure 2. Details of the interaction between the A- and P-domains, viewed from the back side. AMPPCP, Mg2+ (light green) and two water molecules (red) are shown in ball-and-stick. Dotted lines represent likely hydrogen bonds (orange) and co-ordination of Mg2+ (light green). Note that the bound nucleotide and Mg2+ are not well shielded from bulk solvent, suggesting facile exit and entry of ADP and a weak affinity of Mg2+. Supplementary Figure 3. A solvent flattened map at 2 calculated from the initial model containing only the P-domain and M3-M10 helices (shown in atom colour). The part of the atomic model that was not included in the map calculation is shown in orange (N-domain), cyan (AMPPCP), green (Mg2+) and magenta (water). Supplementary Figure 4. A solvent flattened map at 1.5 calculated from the model containing only the cytoplasmic domains and M3-M10 helices (shown in atom colour). The part that was not included in the map calculation (i.e. M1 and M2 helices) appears in orange. Supplementary Figure 5. An initial composite omit map1 (at 2 ; blue net) and Fo – Fc maps (at 5 (pink net) and 9 (magenta net)) around AMPPCP, calculated Toyoshima and Mizutani (2004-03-16597B) Supplementary Figure Legends 2 for the P21 crystal using the model constructed for the C2 crystal. No Mg2+ or water molecules were introduced into the model at this stage. 1. Brünger, A.T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr. 54, 905-921 (1998). Supplementary Movie. Conformation changes in Ca2+-ATPase for the sequence E2 E1·2Ca2+ E1·ATP, based on crystal structures of E2(TG)1, E1·2Ca2+ (ref. 2) and E1·AMPPCP (this work). Intermediate models were generated by morphing using LSQMAN3 and energy minimised with MARBLE4. They were rendered by MOLSCRIPT5 and RASTER3D6. Prepared by N. Miyashita. 1. Toyoshima, C. & Nomura, H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature 418, 605-611 (2002). 2. Toyoshima, C., Nakasako, M., Nomura, H. & Ogawa, H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution. Nature 405, 647-655 (2000). 3. Kleywegt, G. J. Experimental assessment of differences between related protein crystal structures. Acta Crystallogr. D. 55, 1878-1884 (1999). 4. Ikeguchi, M. Partial rigid-body dynamics in NPT, NPAT and NP gamma T ensembles for proteins and membranes. Journal of Computational Chemistry 25, 529-541 (2004). 5. Kraulis, P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946-950 (1991). 6. Merritt, E. A. & Bacon, D. J. Raster3D: Photorealistic molecular graphics. Macromol. Crystallogr. B 277, 505-524 (1997).