Biology and Activity of Restriction Endonucleases
advertisement

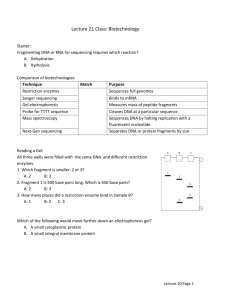
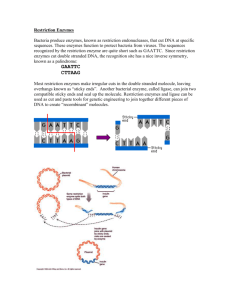
Biology and Activity of Restriction Endonucleases Restriction endonucleases are enzymes that cleave the sugar-phosphate backbone of DNA. In most practical settings, a given enzyme cuts both strands of duplex DNA within a stretch of just a few bases. Several thousand different restriction endonucleases have been isolated, which collectively exhibit a few hundred different sequence (substrate) specificities. Restriction-Modification Systems A large majority of restriction enzymes have been isolated from bacteria, where they appear to serve a host-defense role. The idea is that foreign DNA, for example from an infecting virus, will be chopped up and inactivated ("restricted") within the bacterium by the restriction enzyme. The presence of restriction enzymes immediately begs the question of why they do not chew up the genomic DNA of their host. In almost all cases, a bacterium that makes a particular restriction endonuclease also synthesizes a companion DNA methyltransferase, which methylates the DNA target sequence for that restriction enzyme, thereby protecting it from cleavage. This combination of restriction endonuclease and methylase is referred to as a restriction-modification system. By convention, restriction enzymes are named after their host of origin. For example, EcoRI was isolated from Escherichia coli (strain RY13), Hind II and Hind III from Haemophilus influenzae, and XhoI from Xanthomonas holcicola. Restriction Enzyme Recognition Sequences The substrates for restriction enzymes are more-or-less specific sequences of double-stranded DNA called recognition sequences. Examining the following table will illustrate some important points (recognition sites are shown as double stranded DNA). The length of restriction recognition sites varies: The enzymes EcoRI, SacI and SstI each recognize a 6 base-pair (bp) sequence of DNA, whereas NotI recognizes a sequence 8 bp in length, and the recognition site for Sau3AI is only 4 bp in length. Length of the recognition sequence dictates how frequently the enzyme will cut in a random sequence of DNA. Enzymes with a 6 bp recognition site will cut, on average, every 46 or 4096 bp; a 4 bp recognition site will occur roughly every 256 bp. Different restriction enzymes can have the same recognition site - such enzymes are called isoschizomers: Look at the recognition sites for SacI and SstI - they are identical. In some cases isoschizomers cut identically within their recognition site, but sometimes they do not. Isoschizomers often have different optimum reaction conditions, stabilities and costs, which may influence the decision of which to purchase. Restriction recognitions sites can be unambiguous or ambiguous: The enzyme BamHI recognizes the sequence GGATCC and no others - this is what is meant by unambiguous. In contrast, HinfI recognizes a 5 bp sequence starting with GA, ending in TC, and having any base between (in the table, "N" stands for any nucleotide) - HinfI has an ambiguous recognition site. XhoII also has an ambiguous recognition site: Py stands for pyrimidine (T or C) and Pu for purine (A or G), so XhoII will recognize and cut sequences of AGATCT, AGATCC, GGATCT and GGATCC. The recognition site for one enzyme may contain the restriction site for another: For example, note that a BamHI recognition site contains the recognition site for Sau3AI. Consequently, all BamHI sites will cut with Sau3AI. Similarly, one of the four possible XhoII sites will also be a recognition site for BamHI and all four will cut with Sau3AI. One other point to notice from the table above is that most recognition sequences are palindromes - they read the same forward (5' to 3' on the top strand) and backward (5' to 3' on the bottom strand). Most, but certainly not all recognition sites for commonly-used restriction enzymes are palindromes. Most restriction enzymes bind to their recognition site as dimers (pairs), as depicted for the enzyme PvuII in the figure to the right. Patterns of DNA Cutting by Restriction Enzymes Restriction enzymes hydrolyze the backbone of DNA between deoxyribose and phosphate groups. This leaves a phosphate group on the 5' ends and a hydroxyl on the 3' ends of both strands. A few restriction enzymes will cleave single stranded DNA, although usually at low efficiency. The restriction enzymes most used in molecular biology labs cut within their recognition sites and generate one of three different types of ends. In the diagrams below, the recognition site is boxed in yellow and the cut sites indicated by red triangles. 5' overhangs: The enzyme cuts asymmetrically within the recognition site such that a short singlestranded segment extends from the 5' ends. BamHI cuts in this manner. 3' overhangs: Again, we see asymmetrical cutting within the recognition site, but the result is a singlestranded overhang from the two 3' ends. KpnI cuts in this manner. Blunts: Enzymes that cut at precisely opposite sites in the two strands of DNA generate blunt ends without overhangs. SmaI is an example of an enzyme that generates blunt ends. The 5' or 3' overhangs generated by enzymes that cut asymmetrically are called sticky ends or cohesive ends, because they will readily stick or anneal with their partner by base pairing.