martin1_33696
advertisement

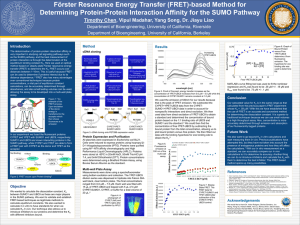
SUPPLEMENTARY DATA: FUNCTIONALITY OF DOUBLY TAGGED SUMO SUPPLEMENTARY FIGURE 1: FIGURE CAPTION: Validation of linearity of signals and non-interference of fluorescent protein tags: The cleavage of EYFP-SUMO1-ECFP (triangles) and wild type SUMO1 (squares) are presented together with the FRET acceptor peak. Densitometric analysis of timepoint samples fractionated on SDS electrophoresis gels was used to calculate the percentage of cleaved protein, presented as a function of incubation time. Fluorescence emission data was collected in parallel and re-scaled as processed protein using the maxima and minima of the 530 nm emission peak. METHOD: In order to measure binding accurately with FRET assays, the fluorescent protein tags must not decrease the functionality of the binding partners. Furthermore, the steadystate binding assay relies on the linear proportionality between the acceptor peak and the number of bound proteins undergoing FRET. These conditions are demonstrated in an experiment that compares the processing of untagged SUMO1 and doubletagged YFP-SUMO1-CFP by a protease, while also monitoring the emission at 530 nm for the tagged reaction. 5 µM wild type SUMO1 and YFP-SUMO1-CFP were incubated with 2 nM SENP1 at 20oC in 50 mM Tris/HCl pH 7.5, 150 mM NaCl, 5 mM -mercaptoethanol. A range of 10 µl timepoint samples were withdrawn and were fractionated by SDS polyacrylamide gel electrophoresis before Coomassie staining and analysis by densitometry (Image Gauge V3.45). Fluorescence was monitored at 530 nm on a Cary Eclipse Spectrophotometer in kinetic mode in intervals of initially 12 s and then 1 min. Data is presented in % of cleaved SUMO. RESULTS: Densitometric analysis of SDS electrophoresis gels shows that the fluorescent fusion protein cleaves at a similar rate to the wild-type SUMO1 and confirms that doubletagging SUMO1 with ECFP and EYFP does not decrease its functionality in this case. The 530 nm data follows the cleavage rate accurately and confirms that this is a proportional measure of the number of molecules undergoing FRET. SUPPLEMENTARY FIGURE 2: FIGURE CAPTION: FRET multi-well plate assay results: Binding curves for EYFP-Ubc9 and ECFPSUMO1, and EYFP-Ubc9 and ECFP-RanBP2. The resulting binding constants are Kd = 0.66 ± 0.11 µM and Kd = 0.17 ± 0.04 µM respectively. SUPPLEMENTARY FIGURE 3: FIGURE CAPTION: Analysis of the affinity of two RanBP2 fragments for SUMO-1 by glutathione affinity chromatography. A lack of RanBP2(l)(2532-2767) bands demonstrates neither SUMO1 nor GST affinity, whereas the RanBP2(s)(2633-2762) band shows SUMO1 affinity. METHOD: 5 M GST or GST-SUMO1 were mixed in 100l with 5 M RanBP2(l)(2532-2767) or RanBP2(s)(2633-2762) buffered in 50 mM Tris/HCl pH 7.5, 5% Glycerol, 5 mM DTT, and incubated at 25C for 1 hour. GST proteins were removed from the mix by addition of 10 ml glutathione agarose beads, which were thoroughly washed before addition of 30 l Laemmli’s sample buffer to elute the GST and associated proteins. 1/2 load and elutions were fractionated by 12% acrylamide SDS polyacrylamide gel electrophoresis before analysis by Coomassie staining.