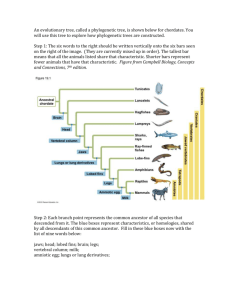
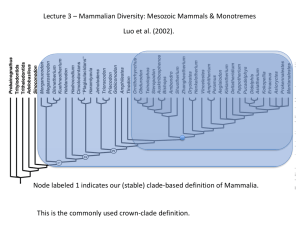
Phylogenetics Reading Assignment 1
advertisement
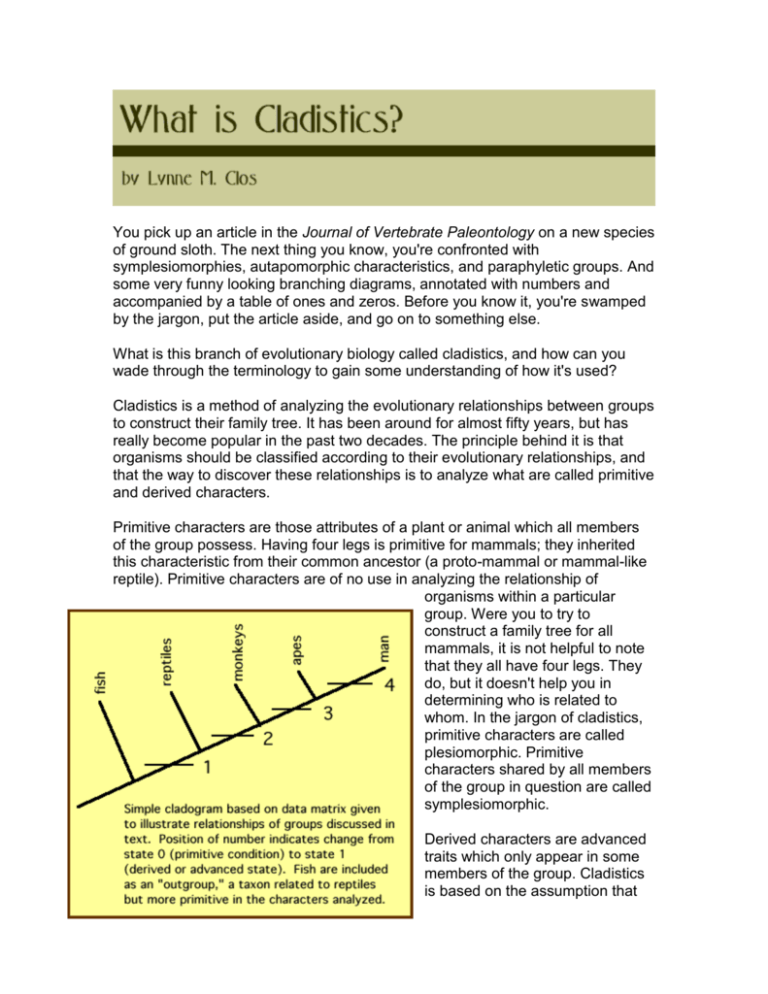
You pick up an article in the Journal of Vertebrate Paleontology on a new species of ground sloth. The next thing you know, you're confronted with symplesiomorphies, autapomorphic characteristics, and paraphyletic groups. And some very funny looking branching diagrams, annotated with numbers and accompanied by a table of ones and zeros. Before you know it, you're swamped by the jargon, put the article aside, and go on to something else. What is this branch of evolutionary biology called cladistics, and how can you wade through the terminology to gain some understanding of how it's used? Cladistics is a method of analyzing the evolutionary relationships between groups to construct their family tree. It has been around for almost fifty years, but has really become popular in the past two decades. The principle behind it is that organisms should be classified according to their evolutionary relationships, and that the way to discover these relationships is to analyze what are called primitive and derived characters. Primitive characters are those attributes of a plant or animal which all members of the group possess. Having four legs is primitive for mammals; they inherited this characteristic from their common ancestor (a proto-mammal or mammal-like reptile). Primitive characters are of no use in analyzing the relationship of organisms within a particular group. Were you to try to construct a family tree for all mammals, it is not helpful to note that they all have four legs. They do, but it doesn't help you in determining who is related to whom. In the jargon of cladistics, primitive characters are called plesiomorphic. Primitive characters shared by all members of the group in question are called symplesiomorphic. Derived characters are advanced traits which only appear in some members of the group. Cladistics is based on the assumption that the appearance of derived characters gives clues to evolutionary relationships. In our example, a derived character for some mammals might be loss of the tail, which occurs in the great apes and man. It is assumed that loss of the tail occurred only once, in the common ancestor of apes and man, and that none of us has one because we inherited that trait from our common ancestor. Thus if mammals are separated into groups which do and which don't have a tail, shown by a fork on the evolutionary diagram (cladogram), this represents the point at which a new species evolved which didn't have a tail. Man and the great apes are assumed to have descended from this species (which may or may not remain undiscovered at the present time). Derived (advanced) characters are called apomorphic in the lingo of cladistics. If they belong only to the group in question, they are called autapomorphic; obligate bipedalism (two-footed walking) is an autapomorphy of hominids (people), a characteristic which we do not share with the great apes. If the derived character serves to unite two groups, it is called synapomorphic; loss of a tail is a synapomorphy of the group containing great apes and man. No animal within the group has a tail, yet our next-nearest relatives, the monkeys, do. It is important to note that the designation of primitive and derived characters has meaning only when related to the group under study. A character which is derived relative to one group may be primitive for a less inclusive group. The occurrence of fur is a derived character if one is studying all tetrapods (four-footed vertebrates), and serves to distinguish mammals from their ancestors, the reptiles. However, it is a primitive character for the group consisting only of all mammals, and is not useful for determining relationships within the Mammalia. The premise behind a cladistic analysis is that by examining suites of primitive and derived characters, diagrams can be drawn which illuminate the evolutionary relationships between the groups. Branching points (nodes) on the diagram are generated every time a derived character (or group of them) is identified which one group possesses and another does not. The two groups on alternate sides of a node are called sister-groups. By analyzing enough different characters or traits, eventually, it is hoped, a true picture of the family tree can be generated. The goal is to create a diagram where all members of the analysis are descended from a single, common ancestral species, and for which all descendant species are included. This is called a monophyletic (or, sometimes, holophyletic) group. If all members of a group are not descended from a single common ancestor, the group is termed polyphyletic. If the group doesn't contain every descendant of that common ancestor, it is called paraphyletic. An example of a paraphyletic group is the reptiles. The Class Reptilia in its traditional sense is a useful concept, but it doesn't contain all the descendants of a common ancestor, because mammals and birds are generally placed in their own classes. (Dyed-in-the-wool cladists are dead set against allowing classifications which contain paraphyletic and polyphyletic groups.) There are some factors which complicate a cladistic analysis. One is convergent evolution. We said above that bipedality was a characteristic of man. But if your analysis included all groups of mammals, you would probably note that kangaroos are also bipedal. Does this mean that they should be considered closer relatives of man than are the great apes? Intuition tells you they should not. This problem is handled by including as many different characters as possible in a cladistic analysis. When one, like this, clearly doesn't fit the pattern of all the others, it is assumed to be an anomaly and overridden in the analysis. Reversals can cause problems, too. We said above that all mammals have fur. Whales don't (to any significant degree), but that is because the fur their mammalian ancestors possessed has been lost in an aquatic environment. Again, when anomalous characters like this are encountered, they are overridden in the analysis. It is important to use enough characters in the study that things like this will shake out. Another problem arises when dealing with fossil material in that sometimes parts of the animal are missing which are needed to evaluate a particular character. In this case, the characters are simply scored as missing (frequently designated by a question mark); they are just ignored (for that specimen) when generating the cladogram. This problem is handled by gathering data on as many characters as possible so that no one deficiency is likely to throw the analysis off. Cladistic analyses are generally run on the computer using the principle of parsimony. This basically means that the computer generates all possible family trees which would fit the data, and you assume that the simplest one is probably correct. To use a computer program such as PAUP or MacClade, you must first convert the results of your study into a data matrix. That's the table of ones and zeros you see in many papers. This is simply a shorthand for, does a particular animal have a certain characteristic, or doesn't it? (Is it bipedal, or not? Does it have a tail, or not?) Although multistate characters are sometimes used, it is most common to split the analysis up into a series of either/or questions which are then scored yes or no, one or zero. If you know which is the primitive and the advanced state, you usually assign zero to the primitive and one to the advanced, but this is not necessary. The computer can handle the unpolarized data matrix and give you the simplest diagram without that information. The more information you include in your analysis -- the more species, and the more characters -- the more likely you are to come close to the "true" family tree. Although cladistics is touted as the greatest thing since sliced bread and a totally objective method, it is still no better than the researcher who decides what characters are important to use in the analysis. It is also no better than the completeness of the fossil material available. The real merit of cladistic methods is in their use of shared derived characters to unite groups and in the ability of the computer to handle large batches of data. Over time, things will change, and new cladograms will be generated. Don't be intimidated by them the next time you try to read a paper in the primary literature. They're really just highfalutin diagrams which try to illuminate evolutionary relationships.