mec13039-sup-0010-AppendixS2
advertisement

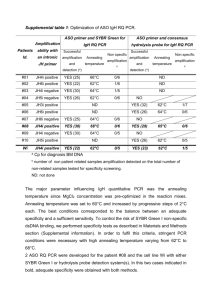
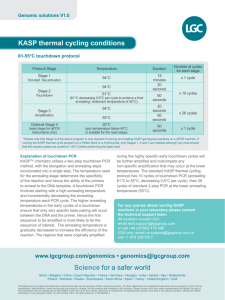
Appendix S2 Amplification and genotyping MHCIIB characterisation We amplified contemporary samples by relying on 23 cycles during the first stage of PCR, and 5 cycles during reconditioning PCR (Lenz & Becker 2008; Sutton et al. 2013). For historical sample DNA, we achieved similar amplification to contemporary samples (as confirmed on agarose gels) with 32 and 36 cycles during the first stage of PCR for robins and saddlebacks respectively (most saddleback samples were older, requiring additional cycles to achieve the same level of amplification). We used 8 cycles during reconditioning PCR. During PCR for historical samples, we were careful to ensure that similar levels of amplification occurred as for contemporary samples, so that unnecessary PCR artifact formation was avoided despite the additional cycles (i.e. we stopped amplifications prior to reaching the plateau PCR stage). For robins, we were able to employ the same PCR conditions as described for saddlebacks in Sutton et al. (2013), except that we decreased the annealing temperature to 65 °C. After next generation sequencing, our MHCIIB dataset consisted of 304 amplicons representing 262 individuals (69 SI saddleback; 69 NI saddleback; 58 SI robin; 66 NI robin; 42 samples replicated) sequenced across three Ion Torrent™ experiments (Life Technologies; Rothberg et al. 2011). After removing low frequency reads (<0.001 of variants), a total of 981,937 reads were assigned to individual amplicons based on exact matches of barcodes on both the forward and reverse primers. The average number of reads per amplicon was 3,230 (± 1,774 SD, range 223 – 12,250). Each saddleback nucleotide sequence included 80 complete codons representing 240 bp of exon 2, while each robin sequence was 228 bp and included 76 complete codons. Average repeatability between replicate samples was 0.97, indicating that a very high proportion of sequences were consistent across replicated samples, and which is similar to other next generation sequencing studies (Sepil et al. 2012; Sutton et al. 2013). Microsatellite amplification and genotyping Our thermocycling conditions for contemporary samples were 95 °C for 5 min, followed by a touchdown sequence comprising 94 °C for 30 s, annealing for 90 s and extension at 72 °C for 45 s. During the touchdown cycles, the annealing temperature started at 60 °C and reduced 1 degree per cycle for 8 cycles. This was followed by 25 cycles at 94 °C for 30 s, 52 °C for 90 s and 72 °C for 45 s with a final 30 min hold at 60 °C. For historical samples, we followed similar protocol, except that we increased the number of touchdown cycles to 12 (starting at 60 °C and ending at 52 °C), and the number of constant annealing temperature cycles to 30. For the historical samples, we also increased the extension time to 60 s. References Lenz TL, Becker S (2008) Simple approach to reduce PCR artefact formation leads to reliable genotyping of MHC and other highly polymorphic loci - Implications for evolutionary analysis. Gene 427, 117-123. Rothberg JM, Hinz W, Rearick TM, et al. (2011) An integrated semiconductor device enabling non-optical genome sequencing. Nature 475, 348-352. Sepil I, Moghadam HK, Huchard E, Sheldon BC (2012) Characterization and 454 pyrosequencing of Major Histocompatibility Complex class I genes in the great tit reveal complexity in a passerine system. BMC Evolutionary Biology 12, 68. Sutton JT, Robertson BC, Grueber CE, Stanton JL, Jamieson IG (2013) Characterization of MHC class II B polymorphism in bottlenecked New Zealand saddlebacks reveals low levels of genetic diversity. Immunogenetics 65, 619-633.