Oral antibiotic treatment induces skin microbiotadysbiosis and
advertisement

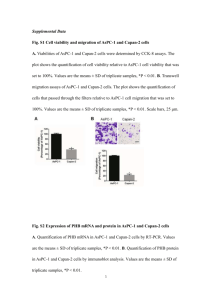
Oral antibiotic treatment induces skin microbiota dysbiosis and influences wound healing Meiling Zhang 1, Ziwei Jiang1, Dongqing Li1, Deming Jiang1, Yelin Wu1, Hongyan Ren2, Hua Peng2, Yuping Lai1* 1. Shanghai Key Laboratory of Regulatory Biology, School of Life Sciences, East China Normal University, Shanghai, People’s Republic of China , 200241 2. Shanghai Majorbio Bio-pharm Technology C.Ltd., Shanghai, People’s Republic of China, 201203. Correspondence: Yuping Lai MinhangDongchuan Road No.500, Shanghai, 200241, China. Email: yplai@bio.ecnu.edu.cn Tel/Fax: 86-21-54342908. Materials and methods Animal treatment and sample collection Wild-type C57BL/6J mice (male, 8-10 weeks) were purchased from the National Rodent Laboratory Animal Resources, Shanghai Branch of China and were housed in the animal facility at East China Normal University. The ECNU Animal Care and Use Committee approved the protocol. To test the influence of a combination of antibiotics, mice were randomly divided into two groups (four mice in each group). One group of mice was given drinking water and the other group was provided with three antibiotics (Vancomycin 1mg/ml, Clindamycin 1.5mg/ml, Polymycin B 1000U/ml) for seven days. At day 6, dorsal skin was shaved after the mice were anesthetized with sodium pentobarbital. The following day, an 8-mm full-thickness excisional wound was made using a biopsy punch and wounds were monitored for another 5 days. At day 12, the mice were euthanized. 2mm of each scar, of the same size and the unwounded skin far away from the scars were collected. PCR amplification and DGGE profiling Skin bacteria were characterized by amplifying theV3 region of the 16S rRNA gene. For amplification, the 25 μL reaction mixture contained 2.5 μL of PCR buffer, 2 μL of 25 mM dNTP mixture, 0.625 U of Ex Taq DNA polymerase and 12.5 pmol of each primer: P2 (5'-ATTACCGCGGCTGCTGG-3') and P3 (5'-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGCCTACGG GAGGCAGCAG-3') as described by Muyzer et al. [1]. The samples were amplified in a thermocycler PCR system by a touchdown PCR protocol: denaturation for 5 min at 94C to denature, annealing for 1 min at 65C. The annealing temperature was decreased by 1C every second during subsequent cycles until touchdown at 55C. Five additional cycles were conducted at 55C. A final extension was conducted at 72C for 3 minutes [1]. Samples from the different groups were loaded onto a denaturing gradient gel electrophoresis (DGGE) gel. DGGE was performed with a Dcode System apparatus (Bio-Rad, Hercules, CA, USA), according to manufacturer’s instructions. Amplification products were separated on 8% (wt/vol) polyacrylamide gels with a linear 35 to 55% denaturing gradient (100% denaturant corresponds to 7 M urea and 40% deionized formamide). Electrophoresis was performed in Tris-acetate-EDTA (TAE) buffer at a constant voltage of 200 V and a temperature of 60C for200 min. The DNA bands were stained with SYBR green I (Amresco, OH, USA) and photographed using a UVI gel documentation system (UVItec, Cambridge, United Kingdom). Real-time quantitative reverse transcription PCR (qRT-PCR) The Trizol Reagent (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA from wounded and unwounded skin samples. The PrimeScriptTM RT regent Kit (Takara, Dalian, China) was used to synthesize cDNA from 1 μg of total RNA, according to the manufacturer’s instructions. Real-time RT-PCR was conducted in a Stratagene MX 3005P (Agilent Technologies, CA, USA). Invitrogen LTD synthesized the primers (Table 1). The comparative ΔΔCT method was used to quantify the gene expression in each sample. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the expression control. All the assays were performed in triplicate and repeated at least twice. Table S1.Primers used for gene quantification in this study Primer Orientation Sequence (5'-3') GAPDH Forward CTTAGCCCCCCTGGCCAAG Reverse TGGTCATGAGCCCTTCCACA Forward TTCCTGTCCTCCATGATCAAAA Reverse CATCCACCTCTGTTGGGTTCA Forward GCTCCAGAAGGCCCTCAGA Reverse CTTTCCCTCCGCATTGACA Forward TTCTCATTGCCCTGTGG Reverse GGCTGCTGGAAGTTGGA Forward TGTTTGAGAAAGGAGGCAGAT Reverse GGAACTTCCACAACTGCCAATC Forward GGCTTCAGTCATGAGGATCCAT Reverse TTTGGGTAAAGGCTGCAAGTG RegIIIγ IL-17A IL-22 mBD3 mBD4 Fig. S1 Impaired wound healing and decreased expression of RegIIIγ mRNA after antibiotic treatment. (a) Photographs of skin wounds after wounding by the biopsy punch at day 12 (The schematic diagram of the experiment is shown in Fig.1a). Antibiotics included polymycin B 1000U/ml, vancomycin 1mg/ml, clindamycin 1.5mg/ml (PVC). Four mice were involved in each group. The area of scar was measured. (b) Quantification of RegIIIγ mRNA expression in unwounded and wounded skin in mice treated with or without antibiotics. n=4. PVC indicates the combination of these three antibiotics Fig. S2 Vancomycin treatment influences the intestinal bacterial structure and decreases the expression of RegIIIγ and IL-17. (a) DGGE pattern of the intestinal bacteria. C, control group; V, vancomycin treated group. (b) Quantification of RegIIIγ mRNA expression in the intestine of mice treated with or without vancomycin. (c) Quantification of IL-17 mRNA expression in the intestine of mice treated with or without vancomycin. *p<0.05,** p<0.01. The p value was analyzed by a two-tailed t-test. Data are the means ± SEM of n=4 Fig. S3 Quantification of the mRNA expression level of IL-22 (a), mBD3 (b) and mBD4 (c) in the unwounded and wounded skin of mice treated with or without vancomycin Reference 1. Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59 (3):695-700


![Historical_politcal_background_(intro)[1]](http://s2.studylib.net/store/data/005222460_1-479b8dcb7799e13bea2e28f4fa4bf82a-300x300.png)