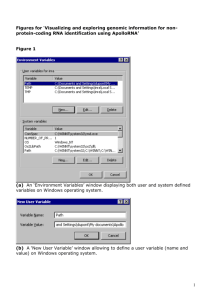
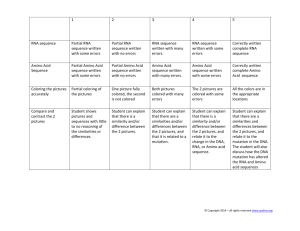
К вопросу о РНК/Белковой
advertisement

Revisited to RNA/Protein symmetry Deichman A. M. (deichman@mtu-net.ru ; amdeich@rambler.ru ) State Unitary Enterprise by N.N. Blokhin Russian Cancer Research Center, Russian Academy of Medical Sciences (SU RCRC RAMS) Introduction Feasibility of some process stages for so-called “reverse translation = rT” was demonstrated for the first time by M.Nashimoto [1]; (familiarity with this article and the book (2005) of author namely this article is advisable). The author synthesized special 83-nucleotide RNA (rtRNAArg), combining methods of state-ofthe-art RNA-technology and RNA-selection in vitro, using T7 RNA polymerase. Such rtRNAArg («reverse translation RNA») simultaneously contained specific hammerhead RNA structure with Arg-binding domain and AGG-codon (Arg) at 3’end. Hammerhead structure contained stem/loop with 3’-end tRNA-similar termination. Usually, the loop included 4 nucleotides (tetraloops, analogue of proteins β-turn) whereas Arg-binding domain (UCCUCACG) contained ССU- and ACG-trinucleotides complementary to Arg codons. It should be noted, that the existence of rtRNA-similar structures among wide-spread different kind of lowmolecular RNAs in cells is not excluded. As it is supposed, hammerhead-ribozyme appeared in epoch of RNA-World (RNA-W; about 3.2 – 3.9 bln years ago) and, probably, is the predecessor for many structural and functional components of present RNA: rRNA, tRNA, etc. Complementary 8-nucleotide (GAGUUCCC) sequence, so-called pre-mRNA, was synthesized in DNA/RNA synthesizer for free 5’-end single-strand portion of rtRNAArg. Using T4 RNA, ligase pre-mRNA was covalently bound by its 5’-end with 3’-AGG-arginin codon. For control purpose both synthesized RNA were provisionally purified by means of gel filtration. Neo-formed 91-nucleotide duplex complex (rtRNAArg/pre-mRNA) under certain conditions (50ºС, 10 mM MgCl2) was exposed to autonotching by С80 -2(ribozyme had only this activity). Then 11-nucleotide mRNA sequence was disengaged from RNA-duplex. Thereby, trinucleotide AGG moved in 3’→5’ direction from rtRNAArg to pre-mRNA. Such 3’→5’ direction is characteristic at least for ribonucleotidic synthesis with U-insertion-deletion RNA-editing in Trypanosoma kinetoplasts, reading RNA-components of telomeres and reverse transcriptases, and pairing nucleotides of tRNA anticodon with nucleotides of mRNA codon. But 3’→5’ directional synthesis on lagging interruptedly replicated RNA-strand is the alternative variant: each of individual RNA fragment is synthesized in conventional 5’→3’ direction and is ligated with adjacent fragment. Since it’s possible to create any rtRNA using the abovementioned methods, the similar experiment can carry out for each amino acid. The author proposed the following: 1) Evolutionary short period of maintaining RNA/protein symmetry prevailed in evolution. This concerned primitive ribosomes when RNA and primitive proteins were inseparably connected. RNA↔Protein-(primitive) conversions were bidirectional, whereas accumulation of RNA-genetic information with rT-mechanism participation was facilitated. 2) rT-mechanism requirement seceded under primarily development of proteinsynthesizing and, later, replicative apparatuses аs well as due to its ineffectiveness and even the danger of genetic overstrain. M.Nashimoto named the difficulties that should be overcome in hypothetical and experimental justifications of rT-mechanism existence in the past. In hypothetical case these are: i) Terminal amino acid of primitive protein should behave like free acid, i.e. to link preferably with its own rtRNA (it should be noted that early evolutional oligofunctional organic/inorganic fixer could be replaced later by polyfunctional bio-membrane). ii) In order to avoid irregularity of rT-process, just terminal amino acid should be firstly cut out from RNA-complex [(N-primitive protein-C)–(3’-AСС-premRNA)]. Secondly, partly degraded protein and partly elongated pre-mRNA 2 -3should be held by the complex (at farther action of polyfunctional apparatus is supposed here also). iii) Each pre-mRNA and each rtRNA, correspondingly, should have common anchor (ССА) and complementary to anchor (GGU) sequences. Obviously the author is exclusively oriented to universal genetic code (modern UGC), although it is unknown whether another universal code or several/multitude predecessor-codes or systems with coding elements were predecessors of present-day UGC (see below). Difficulties to be overcome for experimental justification are connected by the author with necessity to maintain the following: i) Covalent ligation of primitive protein with pre-mRNA (expectative remaining relic of such process is exemplified: RNA-poliovirus is covalently linked with VPg-protein); ii) Exact cutting out only and exclusively C-terminal amino acid (although, activity of multifunctional nucleoprotein complex is possible also); iii) Permanent regeneration of eliminated 3’-codon with participation of singular (autonotching) ribozyme activity (complex structural system action is conceivable also). The author associates potentiality of getting over abovementioned difficulties with development of different RNA-engineering methods. However, even if such solutions are found for each individual case, generally for such a scenario, participation of some still undiagnosed special intracellular multifunctional nucleoprotein machinery is possible. On the analogy with others (ribosome, spliceosome, primosome, editosome) it can be named “retranslosome” (see below). It’s clear that initially excluding existence of such natural machinery elements in assumed RNA/protein symmetry period (and, all the more, at present) M.Nashimoto makes the following conclusions, supposing that: 1) Over evolutionary significant duration bidirectional (RNA↔Protein) RNA/protein symmetry gradually degenerates (reverse-function regresses – rT- 3 -4mechanism disappears) into one-directional (RNA→Protein) movement of biologically valuable information. 2) First primitive, than full-value cellular ribosomes are formed and later – replicative apparatus also. 3) It’s essential to search for relics (for example, structurally-like tRNA and elongation G-factor) of this ancient mechanism. However, in future we’ll search for not only relics, but also rT-like mechanism itself and some present evidences of its activity. Among hypotheses out of “egg/chicken” series (necessary for revelation of nucleic and/or protein biopolymers synthesis antecedence) the following are reviewed by the author: RNA-first (RNA-W), RNP-first, Protein-first, but DNAlatter. Actually, deoxynucleotides are far harder in reproduction under pre-biotic synthesis conditions in comparison with ribonucleotides, are more stable and preferable for genetic information storage. Their biosynthesis (as well as DNAfragments of lagging strand and deoxysugars) in relation to ribonucleotides is secondary. Moreover, RNA synthesis, as judged by speed (50 nucleotides per second against 500–1000 for DNA) and replication error level (10-3–10-4 for RNA- and 10-9 for DNA-copying), is more “primitive” [2]. M.Nasimoto prefers RNA/protein symmetry, i.e. early genetic code formation at RNA/protein parity conditions (RNP-first period). RNA-first theory is considered to be the most developed among mentioned (see “RNA World” 1999, ed. by R.F., Gesteland, T.R., Cech, J.F., Atkins, CSHL-Press). According to this theory and de facto RNA molecules possess informative and many enzymic properties simultaneously. Nevertheless, latter are less expressed than in proteins: structural compromise inconsistency arises because of different requirements to chemical catalysis (dynamic structures priority) and information capacity (conservative structures priority) in the same molecule [3]. Ribozymes and RNA-World theory 4 -5Small simple ribozymes (say, autonotching/ligating hammerhead, hairpins, HDV, VS-retroplasmids of Neurospora fungus mitochondrions, etc) are not so active enzymes as protein ones. The latter are more flexible and abundant in ligands (of amino acid side groups) and dehydrated Me-ion-binding sites. Hydrophobic protein centers are more stable than RNA polar centers (with weakly hydrophobic nucleus) open for intra-molecular interactions even in continuous dielectric medium. Functional activity of the latter is connected with rich inclusions of several irregular structures (stem/loop, hairpin, bulging, one-strand RNA, divergence, etc). These structures deviate from classic A-form helix. Some of them (stem/loop, etc.) are evolutionary oriented to interacting with proteins in RNP composition. Such like ribozymes minimal size is 16–34 nucleotides. For small ribozymes secondary structure as well as for proteins tertiary structure restoration (due to hydrophobic interactions), renaturation proceeds over several microseconds. However, small ribozymes do not need the presence of proteins for required conformation support, whereas proteins need the presence of RNA (inside RNP). At the same time global ribozymes are strongly hydrated even on hidden sites. They are more linear than protein globules and restore super-helical 3Dstructure in several orders of magnitude slower (over milliseconds, seconds and even minutes) and with larger conformation variations. Metal single-ions localization and ribozymes catalytic sites can be different or partly overlapping. Among ions Mg2+ is the basic ion, less frequently occur Mn2+, Ca2+ as well as di- and poly-ion sites [4-7]. Under folding conditions hydrophobic interactions for proteins and Hlinkages, generally in triads of stacking flat bases, for RNA are predominant. A number of folding structural units interactions features proceed with involvement of structural organized water and are determined by the balance between Van der Waals forces (usually repulsing) and electrostatic London forces (usually attraction) having action radius r12 and r-6, correspondingly. The RNA multiple conformational structures is formed by 29 pairs of nucleotides whereof only 2 are Watson-Crick’s. Just those prevail in conformational harder DNA-folding [8]. 5 -6Natural ribozymes in the whole (for example I group introns) are most active in relation to nucleic acids sequences and less to proteins, whereas proteins are most active in relation to low molecular substrates and themselves. In both cases activity limiting components are the transient state stability and reaction product dissociation rate. However, both zymes species use similar catalytic strategies combinations and exhibit other activities, but proteins have still higher catalysis specificity [6]. RNA-W theory is fairly contradictory (F., Crick: “there is the gap between prebiotic soup and RNA-systems), and a number of different RNA-W formation pathways can be found in different authors publications. Theory only partly answers to the question about origin of high-level complexity protein-synthesizing and replicative apparatuses (and accordingly of life). Origin of abiogenous and prebiotic chirally active universal code predecessors is not clear (their number is so high that “prebiotic life nightmare” conception is actually). It is supposed, that nucleotide and even non-nucleotide genetic forming systems could be among them [108]. Origin of the following is also unclear: cis-/trans-mRNA coding (splicing), functional long coding RNA molecules from prebiotical short ribozymes, early non-template biopolymers synthesis [4] and present-day tRNA and Aa-tRNA-synthetases [9]. It’s of interest, that not only tRNA but also small hairpins can be synthetases substrate [10]. Furthermore, period of RNA-W formation on the Earth (especially for initial 500 million years) and priority of single- and dipolymeric biological systems appearance for early evolutionary stage are not clear. In general, obviously, it is naive [108] to think that “ever-young” origin of life problem will be solved within near decades. Moreover, RNA-W can be non-confirmable [6] whereas RNA can be not the first single-polymer catalytic system [3]. That is why it is appropriate to study along with other an approach assuming not only lost (as M. Nashimoto does) but factual mechanism. Besides, it should be taken into account that verifiability of each really existing mechanism is usually much more preferable than of a lost one. The possibility of search for genetic code roots out of RNA-World is questioned 6 -7It is still uncertain just when and out of which predecessor, RNA or DNA [11], LUCA, last universal common ancestor of genetic code, appeared (at least 500 million years ago). Nucleic sequences (i.e. direct evidences), age of those could be put down for RNA-/RNP-/Protein-/DNA-Worlds epoch (billions years ago), have not been found and, probably, will not be found using present methods. The ancient fossils (of different nature inclusive of aged more than 3 bln years), including bacterial, are merely morphologic structures (pseudomorphs). In some cases their nature may be even abiogenous [12]. M. Nashimoto fairly notes (and we partly add) that: 1. Abiotic formation of several (~10) kinds of different biochemical metabolite units could go before universal genetic code based on presently dominating nuclear acids [13]. Or else, several (up to 10; 7 of which are in accordance with the theory of universal code coevolution and biosynthetic pathways of amino acids) evolutionary cascades [14]. Theory reflects the stage for which physicochemical properties of interacting amino acids and corresponding codons are already important. Although, codons, as in Glu/Gln and Asp/Asn cases, can be transferred from pre-aminoacids to final amino acids without changing biosynthesis pathway; 2. Primitive protocell contained relatively small sized RNA and proteins molecules; 3. Each of hypotheses is considerably contradictory. (3a) Theory Protein-first assumes self-replication not an individual molecule, as in case of RNA-replicase of RNA-W epoch, but proteinoid complexes. Probably, they reproduced on thermal polymerization basis (such kinds of activity: phosphatolysis, esterolysis, decarboxylation, deamination, oxidation and photochemical decarboxylation, etc.) still before cell division. Notice that by now some activities, important for self-replication of peptides groups (but non individual ones), are virtually shown [15-17]. In 14 amino acids peptides can by factor of 106 intensify activity of ribozymes tenfold larger in size [3]. 7 -8(3b, 3c) Whereas in RNA-/RNP-first theories ribozyme auto-replication is possible only for considerably long (at least 300-600 ribonucleotides) molecules, the way of emergence which is fuzzily. It is even conceded that RNA-similar selfreplicating polymers existed before RNA-W epoch [18]. 4. Formation of translational apparatus and genetic code is little-known; 5. Actual rT- (or like) mechanism may greatly differ from the proposed one. In connection with such mechanism, new widely used biotechnological approach may appear (in proteomics, molecular biology, genomics, pharmaco-genomics, gene-informatics, metabolomics and many others). The situation may be even more complicated if we take into account increasing tendencies in development of studied in-vitro/in-vivo natural and artificial DNA-zymes, chimerical DNA-RNA-zymes and their substrates. By now a significant number of experimental works appeared where a variety of specific DNA-zymic activities are shown. Among them are DNA- and RNA-ligasic, DNA- and RNAnotching, DNA- and RNA- nucleasic, DNA-phosphorylating and adenyling, metallating (Н+ is substituted by Ме2+) and others. Activity of certain DNA-zymes is comparable with the same for ribozymes and proteins [19]. On the first hand, these activities are able to affect chemistry of some intracellular processes and individual gene expression. Secondly, determination of these activities has increasingly widening application in different fields: from biotechnology to therapy [19-20]. DNA-zymes activity, as well as of some ribozymes, can be induced by light [21-22] and more expressed in relation to short oligonucleotide RNA-fragments [3, 18]. So, I group intron containing SynY-ribozyme (~180 nucleotides) Tetrahymena preferably recognizes short (including triplet) helical oligonucleotides [18], copulates with them and multiple reunite them (error level not above 1%). This ribozyme can assemble autocomplementary strand from 18 fragments (~ 10 nucleotides in each). All abovementioned leads to qualitative rethinking of the role of not only DNA and DNA-World theory in evolution processes but also other hypotheses from “egg/chicken” series. Each of them creates own mechanisms, as a rule, with participation of standard nucleotides and amino acids, i.e. is exclusively oriented to 8 -9present universal code. In doing so it is not taken into account that there could be several codes and proposed theoretical constructions, although nor fruitless, but supposedly premature for making conclusions concerning “egg/chicken” interrelation. We do not know and have no justification to think that we will know soon from which genetic systems (or systems with coding elements) should real counting started. Notice only that we would not look for causes giving direct or indirect evidence of possible several codes existence but for ourselves formulated hypothetical mechanism, one function of which can be intergenomic and/or intercode retranslation [23-26]. Abovementioned hypotheses, including the attractive RNA-W, are contradictory. Probably, that is why hypotheses appear in which, particularly, processes of simultaneous DNA-replication/translation ([27-28] and RNAreplication/translation [29] are simulated. Initial (before universal code formation) interaction of amino acids and nucleic acids (copolymerization) is supposed here. The main but not exclusive contradiction in DNA-replication/translation hypothesis is the following: actuation of DNA-self-replicating system formed by one/several aminoacetyl-trinucleotide progenes should be preceded by appearance of hypothetical enzyme, progenligase. That is the protein containing 60–80 amino acids, 15-20Å in diameter which should be able to control at least 8 (!) highly specialized functions for copying coding him nucleotide sequence (similar analogues are not known). However, most probably multifunctional (cellular/protocellular) complex is required in this case. These authors also work within universal code framework pointing at code features giving evidence of possible primarily appearance more simple than present UGC codes. Those are: 1) Expressed duplet (no triplet) coding: physicochemical interaction of amino acids with first two nucleotides of DNA-codons or RNA-anticodons. In this regard two codes are formed: pre-enzymatic (pre-biotic, including about a half of standard and several non-standard amino acids) and modern enzymatic code. 9 - 10 2) Role-defining category of codon’s central nucleotide (relic of pre-duplet coding existence?). 3) Existence of less hard third wobbly position in mRNA codons and in first position of tRNA anticodons. According to opinion of C.R., Woese [30], it is naive to believe that physicochemical correspondence in amino-acid/codone pairing (CAP) even on early evolution stages resolved exclusively within the framework of principle “all or nothing”. Additional weak interactions could act as an important part also. It is especially significant for present coding when powerful multi-component ribosomal “pad” (including rRNA, tRNA, protein factors, etc.) is placed between amino acid and mRNA codon. Not known, whether several codes could be So, all enumerated hypothesizes suppose that nothing but universal genetical code (UGC) should be under consideration, although direct evidences of this are not available. Concerning discussed by M.Nashimoto theory of exterrestrial (Panspermia) bringing-in of life (some nucleotides, amino acids, etc.) out of space [31], it seems effective only in the presence of many other locally necessary evolution conditions, and i.e. it does not solve the problem. Also notice that for any mode of cell division not only nucleic acids but other intracellular components, organelles, complex structures are obligatory inherited. Without finding out of new evolutionary effective special mechanisms place still remains for supporting vulgar-creation viewpoints. Nevertheless, if such mechanism as “reverse translation” (the term is just conditional, from below it’s evident that it’s not correct) is under consideration it’s necessary to discuss a number of problems. It’s essential to analyze possible connection of the mechanism with other known intracellular genome expression mechanisms, molecular biology central dogma (MBCD) and modern scientific 10 - 11 paradigm. It’s also necessary to consider where and how such mechanism can proceed in a cell, why its (or the like) existence in a cell is, most probably, unavoidable. Inevitability existence the similar mechanism will be coordinated to the cybernetic approach within framework of which hierarchy managing and controlled systems and subsystems various levels of organization of cells and organisms it should be supplemented with corresponding feedbacks [113, 114]. Herewith, it is necessary to understand that many code properties (including various stages of genome expression) also are integral parts of modern coding method. Such properties are obligatory connected with genetic code conception. Then, contextually to abovementioned, we’ll name some dominant causes and properties of the code, in connection with which genetic code could evolve, whereas number of codes could be more than one. Such codes (with all their properties) could possibly exist simultaneously, successively, could interact and compete, whereas number of nucleotides in codon as well as nucleotide and amino acid compositions of biopolymers could vary. These causes and properties are the following: 1) Code degeneracy (one amino acid – several codons, origin each whereof could belong to differently coding systems); 2) Prevailing role of the first two (probability of duplet coding) or central (possibility of fore-duplet coding) and decreased role of third nucleotides codon (in tRNA-anticodon it is the first); 3) Utilization of same biosynthetic pathways of aminoacetylation for some different, although related pairs, say Glu/Gln, Asp/Asn, Secis/Ser amino acids (extension of genetic alphabet is possible). 4) Inclusion of great number non-standard nucleotides (over 3 tens for RNA-, and not less than 6 variously CH3-modified their forms for DNA-structures), use of non-standard modes of their pairing (first of all for RNA), various frequency of codone occurrence in genes/genomes (genetic alphabet extension and, possibly, continuing competitive regulating participation of relic forms of nucleotides metabolism); 11 - 12 5) Existence in a cell a wide variety (over 200 accounting β-, γ-, δ-, and εvariants) of natural substandard non-coding amino acids. Some of them, not excluded, could have polymerized properties inside early-evolutionary pre-genetic [26] rather than present genetic systems. Furthermore, “new” amino acids are able to mRNA-context-depending push out stop-codon. These are UGA-coding selenocysteine (Secis; synthesized on tRNA-binded serine), UAG-coding pyrrolisine (Pyl) and selenomethionine (Semet; supposedly UAA-coding). However, possible role of RNA-editing (recoding mechanisms) is underexplored here. Moreover, it’s impossible to ignore that rarely involved (less than 5–10% amino acids) generally are not taken into account by proteinaceous chemists and thrown off as artifact. Worded in items 4 and 5 does not exclude possibility of potential evolutionary dynamic for coding code components; 6) For some reason modern coding does not go without some at first view resource-spending (concerning energy, reproduction of biopolymer components of complicated complexes, time) molecular and cellular, in point of fact searching, processes. Searching is connected with mechanisms of genome flexibility and cellular metabolism. Among molecular processes these particularly are: constant editing of many newly synthesized cellular and virus RNA-transcripts; different kinds of splicing, post-translation modifications, non-triplet translocations, shift/overlapping reading frames, etc. These, particularly, are positive and negative lymphocyte selections amidst cellular processes, when are reproduced unplanned by genome separate nucleotides for synthesis of Ag-specific sites of receptors Тand B-cells; 7) Presence of single- and di-nucleotide coenzymes (NAD, FAD, etc.) as well as specific di-nucleotide preferences for some splicing enzymes [32]. The same is also true concerning conventional and RNA-editing cytidindeaminases [33] for choice of recognizer and target site (hot spots) for RNA- and DNA-modifications. Furthermore, there is an opinion about precedent existence of triplet codes based on one (A), two (A,G), three (A,G,C) and four (A,G,C,U) letters [34]; 12 - 13 8) Existence of semiautonomous monophyletic (for certain taxonomic groups) or non-monophyletic (for other groups), as well as their tRNA [35], cellular organelles. Latter contains own replicating and protein synthesizing apparatuses and, probably, have endosymbiotic origin from α-proteo-bacteria and cyanobacterial progenitors in case of mitochondria and chloroplasts, correspondingly [36]. The list is not complete. In accordance with Orgell’s principle of continuity [108] interacting molecules, their parts and functions clearly or latent co-evolve in a cell [37]. In the judgment of M.Nashimoto, “reverse translation” supported biological continuity in the past. In this regard, a pool was preserved only for those RNA which corresponded just to claimed amino acids sequences. However, it would be premature to call such sequences “proteins”, even primitive, as we do not know their composition and which genetic systems they belonged to. Our approach assumes that the RNA/protein symmetry between nucleic and amino-acid sequences could not to disappear but to be taken over the control of a cell to support such biological continuity. While surviving, such symmetry could support a great number of concurrently developing and interdependent initial pre-genotypic and pre- phenotypic processes. These processes could begin in epoch of abiogenously demanded oligostructures of both types and can be in progress till now. Pathways for genesis of oligostructures could be different: as a result of electrical discharge, prebiotic СО2-fixation on the surface of pyritic crystals, interaction with clay, and others [6]. Another (modern) variant of RNA/Protein symmetry It’s possible to suppose the existence of a mechanism [23, 26] in cell organelles (mitochondria, chloroplasts; more detailed below) up to present for which protein fragment ~ 5-10 fold amino acids (epitope) orients combination tRNA nearby itself (Fig. 1) or aminoacyl-tRNA, Aa-tRNA (Fig. 2). For this case it is supposed that nucleotides of anticodons, at least 3 back-to-back interacting tRNA [38], approach 13 - 14 each other and form helical mini-template for polymerizing reaction (RNAdepending-RNA-polymerase, etc). It is known, that three convex anticodon nucleotide are inside-out and each of them is recognized by 5–6 amino acids AatRNA-synthetase [39-40]. There are nonstandard ones (such as pseudo-uridine, ψ) among nucleotides of anticodones tRNA animal and fungus mitochondria [41]. It is also impossible to exclude participation of coding nucleotides adjacent to anticodon as well as non-coding ones, formed as a result of tRNA-modifying (in the time of RNA-editing) action of anticodone AI-intron [42]. For another variant (irrespective of model choice Fig. 1 or Fig. 2), not excluded, anticodone regions can be cut out and cross-linked again (“primitive splicing”). Necessary for this purpose ligasic, endo-/exonucleasic, proteases and other activities become apparent by RNA-editing and translation/replication/reparation in DNA-containing organelles (mitochondria, chloroplasts) of some organisms. Finally, as a consequence of error, as well as a result of code degeneracy, and non-invariance of point (1-1’, 2-2’,…, 7-7’) interactions of amino acids, amino acid of epitope can be linked with more than one tRNA (Fig. 1) or Aa-tRNA (Fig. 2). Unfortunately, non one of the proposed variants of vIERT-model of NE formation (or even pre-mRNA M. Nashimoto in a context of a given hypothesis) can not be discarded now. Especially, it is in conformity to diatropical principle of multiplicity pathways of achievement the same result [43], in given case concerning of huge number of evolved and variously metabolized of genetic systems. In terms of conformation, it is important that amino-acceptor-type CCA-ends of some 3’-cut-down upstream tRNA from mono-cistronic transcripts of overlapping mitochondria tRNA-genes are subjected to dynamic posttranscriptional transformations [44]. Meanwhile, as a result of alternate degradation and elongation acts length of fragment with CCA-end (per 3–6 nucleotides) and included nucleotides spectrum (CMP>AMP>UMP>GMP) of different animals are varied. However, protection of 3’-end tRNA part from exonucleases is much more effective relating to multiply prevailed canonic CCA-ends. Such protection is provided by aminoacylation, whereas fragment integrity restoration is provided by elongation with 14 - 15 - 4 3 5 2 6 1 7 ССА-stems Аа-tRNA Set L-figurative Аа-tRNA Inner Membrane Organelles 1. 2. 3. 4. 5. Anticodones helical sites Аа-tRNA Polymerase activity Nucleic Equivalent (NE) of Epitope from several codones Fig. 1 кодонов. The variable Individual Epitope Reverse Translation (vIERT) at the adjacent Aa-tRNA anticodon sequences. 1,2,3, … , 7 – amino acid residues of the epitope. ↯ – peptidase activity (perhaps analogical one are and for Fig. 2) 15 - 16 3 4 5 6 2 1 7 6’ 2’ 1’ 7’ CCA-stems Аа-tRNA Set L-figurative Аа-tRNA 1. Inner Membrane Organellas Anticodones helical sites Аа-tRNA Polymerase activity Nucleic Equivalent (NE) of Epitope from several codones Fig. 2 The variable Individual Epitope Reverse Translation (vIERT) at the adjacent Aa-tRNA anticodon sequences. 1,2,3, … , 7 – amino acid residues of the epitope 1’, 2’, 3’, … , 7’ – amino acid residues of the correspondent Aa-tRNA complexes. 16 - 17 - tRNA-nucleotidyltransferases (as well as by poly-adenilisation and, possibly, RNAediting) for eukaryotes, many eubacteries and some archebacteria [44]. Integrity preservation of CCA-endings, potentially able complementary pair with two phylogenetically conservative UGG from 23S rRNA (E. coli) is important for ribosome’s operating in peptidyltransferase center and co-evolving, and tRNA [45]. Abovementioned features of proposed model can promote formation more than one variant of epitope’s Nucleic Equivalent (NE) of ~ 15–30 nucleotides. Among such NEs, in particular, can be sense- and antisens variants. In addition, NE, or even just anticodon juxtaposed parts of Aa-tRNA, not excluded, can serve as peculiar primer for polymerase reaction. Such NE forming mechanism can be conditionally called variable Individual Epitope Reverse Translation (vIERT). For the case of protein synthesis helical mRNA-template length in codonanticodon duplex region (mRNA-tRNA) does not exceed 6–9 nucleotides [9, 38]. This corresponds to two-three back-to-back oriented tRNA. In case of hypothetical retranslosome dimension of such mRNA-tRNA continuum is not known. Holding mechanism for epitope oriented tRNAs (Aa-tRNAs) in mitochondria and chloroplasts [46] is known only partly (see below). Irrespective of NE formation pathway (on juxtaposed anticodons as a template or by “primitive splicing”) polymerization of each one-three neighboring nucleotides into integrated NE, possibly, proceeds with pausing (specific for some RNA-polymerases). For protein synthesis distortion of codon/anticodon interaction in ribosome A-site by different small ligands (medicines, antibiotics, peptides, RNA-aptamers and others) may lead to minor stria displacement of 2-3Å and wrong reading [47]. Similar hindrances are also possible for NE synthesis as a result of vIERT. Presumably, this (vIERT) mechanism is possible at least in mitochondria and chloroplasts (thylakoids). Not external easily permeable for large size (up to 100 kD) particles but internal organelle membranes hardly permeable even for small ions, fix inseparable even in hard lysis conditions membrane-bonded tRNA-fractions [46]. 17 - 18 Distance between interior and external by membranes here can reach more 100Å: that is comparable to the maximal sizes tRNA. Probably, hypothetical vIERTmechanism in organelles, similar to protein synthesizing and replicative apparatuses, is membrane-dependable and can function, as it is supposed, with participation of complex retranslosome. Whereas simultaneous membrane-dependence of protein and nucleic acids synthesis processes as well as hypothetic vIERT-mechanism in organelles, presumably, enables admission of their conjugation [23, 26]. Cell organelles are semi-autonomous structures comprising all necessary activities of replicative, protein-synthesizing and RNA-editing apparatuses. Additional activities supposedly can be provided there by abovementioned RNA- and DNA-zymes. Hypothetical vIERT-mechanism also is not being excluded for nucleus, since not only replication and transcription are shown but tendency of some proteins possible translation has been defined. This concerns, at least, shortened form (M246) A→I of editing enzyme ADAR1, active in relation to transcripts of such proteintargets as GluRs, 5-HT2CR, HDV-antigen and others [48]. This enzyme localizes on nucleolus surface and acts important role in ribosome maturation and in antivirus protection of cell. So, ADAR1 enzyme functions turn out to be intriguingly connected not only with RNA-editing but with its intranuclear translation [48]. For unicellular vIERT-like mechanism can be connected with cytoplasmic membrane (not considered). For cell organelles of different organisms, processes of RNA-editing, provided with different activities combination, are observed. These are ligase, endo- and exonuclease, deaminase (C→U or A→I), uridintransferase, gelicase and other editosome activities. Special RNA-editing modes are observed in nucleus and cytoplasm, but strategy and tactics of RNA-modifications introduction are highly individual for each editing mechanism. Each tissue of organism has individual metabolites level, ATP, GTP and synthesizes particular set of proteins (epitopes). That is why forming or preexisting sets of tRNAs (Aa-tRNAs) in hypothetical organelles retranslosome can be out of chaotic. 18 - 19 Analysis of known RNA-editing modes demonstrated that the process is directly or indirectly dependent on template-information component, whereas transcript fragments – so-called cassettes of 14-29 nucleotides, becomes minimally edited regions [33, 49]. Cassettes size is comparable with such for NE; (it is interesting, that for such heterogeneous objects as the minimum size small ribozymes – 16-34 nucleotides, miRNAs, DNA-fragments in bones and teeth of fossil exhibits, and primers, useable in polymerase chain reaction, is similar also). The following are reckoned among such kinds of RNA-editing: guide-RNA-(gRNA)-dependable Uinsertion-deletion editing in mitochondria of Trypanosomes; dependable on cellular single-/double-strand RNA (ssRNA/dsRNA), cellular and virus exon-intron dsRNAstructures, correspondingly for С→U in cytoplasm and A→I deamination in nucleus and cytoplasm. Protein fragments (epitopes) compel attention of immunologists, molecular biologists, biochemists, virologists, evolutionists and others. Since complex structure and assembly of minimal genetic unit (consisting of many separate parts: promoter, enhancer, exons, introns, terminal sequences, and having individual evolutionary history 5’- and 3’-parts) became known the gene was split. A smaller (similar NE) structure may turn out to be the new minimal genetic unit. Let’s consider in short two cases of hypothetical vIERT-mechanism application for cell organelles (mitochondria and chloroplasts). Some aspects of this question are also represented in materials [26] more widely. vIERT/VLNS-transfer mechanisms in mitochondries of macrophages (APK) It is known that during the foreign antigen (Ag) processing in macrophage (with participation of phagolysosomes, proteosomes, etc) Ag is cut up into separate fragments, linear and conformational epitopes. The latter, not excluded, may be fixed by different cross-linkages after damaging effect of radicals and oxygen toxic products (under oxygen explosion). Among fragments at least three conditional types can exist: similar to own (1), foreign known (2) and foreign unknown (3). In first two 19 - 20 cases antigen-presenting cell (macrophage, dendrite cell, another APC) cooperate the interactions with in various degree activated T- and B-lymphocytes (including memory lymphocytes). Meanwhile, on APC surface Ag fragments are presented together with MHC antigens of I/II class. Thereby, usually one/few already formed units are strengthened or weakened in development of humoral or cellular immune responses of the whole anti-idiotypical chain. Nevertheless, complete specific primary responses (with appearance of new lymphocyte B- and/or T-clones) originate only in third case (new unknown foreign epitope). This variant, not excluded, needs another scenario with the participation of hypothetical vIERT-mechanism. Supposedly, the response can appear not only to qualitative but quantitative (i.e. as the reply to first two fragment types too) disbalance of epitopes subjected to membrane-depending sorting in hypothetical retranslosome (Fig 1, 2). Foreign Ag (epitope), probably, holds on internal membrane of macrophage (Mcrph) mitochondria and provokes energetic and biochemical failure (level/turnover are decreased for ATP, GTP and are increased for radicals and reactive oxygen species). Meanwhile, proton and electron transports may be disturbed in mitochondria normally asynchronous functioning, but in this conditionally pathological situation synchronous [50]. Fixation of such an epitope on internal membrane may initiate vIERT-mechanism (“protective” variant) accompanied by deletion of epitope and reproduction of several NE variations. Newly synthesized NEs, probably, are built in special Vector-Like Nucleic Sequences (VLNS of transposon-/retroposon-like type) for succeeding VLNS-transfer (type of horizontal transfer; possible physiological role of RNP/DNP with NE inside is not discussed) between DNA-containing organelles and cells. If normal immunogenesis includes vIERT/VLNS-transfer mechanisms they can be used for generation of hypervariability mutually evaluating viruses (in particular, HIV; not consider). It is known, that cellular organelles membranes are permeable for some proteins and nucleic sequences. Analogues of intracellular and intercellular (inside of one and between different organisms) VLNS-transfer mechanism are shown for 20 - 21 different genetic systems. This is an exchange of sexual cassettes (plasmids) by yeasts and bacteria. Some viruses, such as rhabdoviridae, bunyaviridae and potyviridae shuttle between photosynthetic and non-photosynthetic organisms exhibiting species- and/or tissue-specificity relating to obligatory host, plants or insects [51]. The role of mitochondrial plasmids is known for nucleus activity regulation in forming the cytoplasm male sterility (CMS) of higher plants [52]. Drug and herbicide resistance in animal cells is connected with maternally mitochondrial plasmids transfer preferably. Natural transport is shown for several tRNA of nonsynthesized in mitochondrions trypanosomes, infecting wide range of plants, invertebrate and vertebrate animals [49, 53]. Radio-autography was used to show RNA flow from trophocytes to Drosophila egg [54], etc. Moreover, for some nucleic acids sequences and viruses, high probability of trans-membrane transfer through mitochondrial pores as a result of experimental electrically induced impulse breakdown is shown. It is considered that such transfer is necessary for inter-mitochondrial and nucleus-mitochondrial exchange in connection with processes related to aging, apoptosis, cellular proliferation, mitochondrial diseases, multiple drug resistance, intracellular particles transport, genome reparation and parental (usually maternal, rarer paternal) mitochondrial heredity [55-56]. It is supposed that hypervariability formation in antibodies and Ag-specific regions of B- and T-cells receptors (BCR, TCR) proceeds with participation not only recombination V-(D)-J-C processes but, possibly, associated to RNA-editing and hypothetical vIERT/VLNS-transfer mechanisms. At that, lymphocytes undergo differentiation (for B-cells it is lymphopoiesis and immunogenesis, including somatic hyper-mutations, SHM, and class-switching recombinations, CSR). Changes in rearranged genes of both receptors [57] are associated, inclusive of, with incorporation of several joining, inter-segment non-coding nucleotides (of P- and Ntypes). Hypervariability (i.e. process of programmed or randomly programmed enumeration of variants) can be formed with the participation of vIERT-mechanism. 21 - 22 Such a mechanism, presumably, takes part not only in generation but in limitation of excess variability at Ag-specific receptor regions. Actually, number of potentially possible receptor variants (up to 10 16 for B- and 1018 for T-lymphocytes) by several orders of magnitude exceeds the total number of lymphocytes in animal organism [57,109]. This variant, particularly, is affordable if nature of P- and Njoining nucleotides appearance is connected with reproduction of NEs or RNA editing which in turn depends on reproduction of NEs (peculiar guiding minimatrixes) obtained as a result of vIERT/VLNS-transfer mechanisms exploitation (described below). It is known, that complete specific immune response requires close physical contact between Mcrph and Т-helper, and Т-helper and Low-Differentiated Precursor of hematopoietic cell (LDPhc). That is why, it is supposed [23, 58, 59], that transfer VLNS with NE inside (Fig. 3) is carried out in the range Mcrph→Т-helper→LDPhc (in bone marrow and, possibly, in thymus). Whole groups of cells (among those various APC, T-cells, B-cells, epithelial, NK, target-cells, etc) come to close physical contact by pairs. These cells participate in such immunological (and not only) important processes as apoptosis, activation, proliferation, positive/negative selection, lymphocyte differentiation as well as under some adhesive interactions and cytolysis [109]. Among plurality of synthesized NEs only one/few participate in the process of positive/negative selection of lymphocytes, on by 95%-99% subjected to death. Herewith qualitative and quantitative characteristics of NE transferred to LDP during VLNS-transfer may play a role of primary anti-apoptotic signals (and formation of the certain ratio between others hemopoietic cells too). It is of interest, firstly, the existence of vIERT/VLNS-transfer-like mechanisms for the case of attitude to central dogma alterations was predicted by F.M.Burnet, the author of clonally selection theory in early sixties [60]. He also considered unlikely pre-existence of absolutely all information necessary for immune response. Another question is that pre-existence of special response-forming mechanisms is possible, but not ready final result. Secondly, it should be noted that vIERT-mechanism (1) 22 - 23 does not conflict with central dogma, because not a whole protein is considered but only its small fragment having conformations combination differing from such in a VLNS (VLNS-transfer) Аg (Foreign Ag) 1. 2.Mcrph 3. 4. 5. 6. Т-help T-help LDP Area of tight physical contact. Fig. 3 Hypothetical vIERT/VLNS-transfer mechanisms possible responsible for hypervariability in Ag-specific areas immunoglobulin molecules. Ag – antigen; Mcrph. – macrophage; T-help. – T-helper cell; LDP – Low-Differentiated Precursor in bone marrow (and possible in thymus). whole protein composition. Moreover (2), this mechanism assumes not invariant (as in case DNA↔RNA→protein) but variable method decoded information reading. But then, apparently, protein epitopes and their NEs from one side (rT-mechanism; 23 - 24 components size – nanomolecular), as well as whole (high-molecular) proteins and genes from the other side (for translation) are elements of different-level systems with non-identical properties. Supposedly, it may be connected with some not disclosed as yet internal specifics of present universal code genesis and functioning [26]. Historically, scientific works of certain authors [61-65] are concerned with «reverse translation» problem at least; probably, it is a theme of separate paper. Only one from these authors [64, 65] employs this mechanism in relation to protein epitope. However, in all these works possible connection of hypothetical mechanism with other genome expression mechanisms is not proved or even predicted (and that was quite natural for that period of time). The same is also true in relation to localization of mechanism in a cell and its general non-contradictory incorporation into the entire paradigm (if it possible). vIERT/VLNS-transfer mechanisms and higher plants chloroplasts Application of vIERT-mechanism for mitochondria and chloroplasts can be different in spite of some features of structural-functional, genetic and metabolic similarity as well as community of organelles origin. Actually, the differences may be connected at least with the fact that not more than almost strictly one tRNA for each amino acid is found in mitochondria: for animals (2-23), fungi (7-26) and plants (2227) tRNA genes. At the same time the numbers of species (30-33) and tRNA-genes (37) are in correspondence with the same for nuclear-coded and functioning in cytoplasm tRNAs in chloroplasts [41, 66]. Moreover, majority of genes in mitochondria have nuclear twins, whereas in chloroplasts up to one third of genes are uniquely coded. Besides, there are many codons in mitochondria and few in chloroplasts which have non-collinear to universal coding sense or are rewritten by RNA-editing at transcriptional level. RNA editing is observed in chloroplasts much rarely than in other cell compartments. Perhaps, it is connected with the fact that after endosymbiotic events different kinds of nucleotide changes appeared and disappeared 24 - 25 here even much faster [26]. Finally, only chloroplasts contain light-absorbing antenna complex of its third (thylakoid) membrane. Light can initiate splicing (cutting-out of I and II groups introns) and transcription of some genes (such as photosystem-2 psbA gene; without light nonspliced transcript is accumulated) and regulate certain cyclic process in cells and whole organisms [23, 59, 67]. Analysis data for nucleotide sequences of all three organelles homological genes enable supposition that genetic information evolutionary movement proceeds from chloroplasts to mitochondrions and nucleus but not in the opposite direction [68]. Causes of such directivity remain unknown, but may be explain in context our mechanisms (see below). It is supposed that application of vIERT/VLNS-transfer mechanisms in chloroplasts may be the following. Scheme 1 shows process of conformity formation between amino acids and nucleotides NE of particular epitope (Fig. 1, and Fig. 2) inside of hypothetical retranslosome. Such conformity on chloroplast internal membrane (in thylakoid grains region), supposedly, may form as affected by total Energy Ray Flow (ERF; first of all it concerns flows with differing photons combinations energy level). This proceeds against the background of all relatively strong and weak field as well as physicochemical specifics of given Earth surface region (biosphere). Simultaneously this conformity may reflect processes of both universal genetic code formation (modern UGC) and diversity within its framework, adequate to concrete evolution stage context. ERF is composed of solar (most powerful), earth (background radiation) and space (weak but evolutionary continuous radiation) components. For photosynthetic organisms ERF itself evolves (unrepeatable by its spatiotemporal characteristics) in two ways. On the first hand, in connection with absolute (acyclic) astrophysical component: the disposition of stars, galaxies and other space objects altered relative each other, at least in the visible part of the universe. On the second hand, in connection with locally-geographical (including cyclic) component: roughly, 25 - 26 - Total Energy-Ray Flow (ERF) against the background of all field and physico-chemical peculiarities of the given Earth surface Amino acids of protein epitope Nucleic Equivalent of protein epitope region (the biosphere) Scheme 1 Formation of amino-nucleic epitope accordance Formation of amino-nucleic epitope accordance (namely the accordance between the amino acid residues of epitope and the nucleotides of nucleic equivalent, NE, epitope) in the Universal Genetic Code (UGC) under the influence of total Energy-Ray-Flow (ERF) including solar, terrestrial and cosmic components, and against the background of all field and physicochemical peculiarities of the given Earth surface region (the biosphere) in hypothetical “retranslosome” chloroplasts of photosynthesizing organisms. 26 - 27 combinations of photons potentially absorbable by photosynthetic organisms, for example, at the Equator and nearby earth's Poles, obviously are not equal. The vIERT-mechanism in chloroplasts (Scheme 1), supposedly, may act as “the retranslator of special kind” [24, 69]. Retranslation may be connected with translation and adaptation, firstly, “elementary particles language”, ERF components [70]. First of all it concerns photons combinations (differing by energy levels; other particles are not considered), supposedly, are variously absorbing by different ingredients of membrane components of light-absorbing structures of various photosynthetic organisms. Only, metabolism of these glico-lipo-proteid components, generally (but not completely), clearly, is genetically determinated. Hereafter, secondly, translation from “elementary particles language” can be carried out by means of quickly-operating “elementary quasi-particles language” application on the surface of chloroplasts liquid-crystal membrane structures chloroplast (thylakoids). Among such biophysical quasi-particles, previously described for solid body, are the following: phonon, polariton, magnon, exciton, soliton, etc [71-72]. Quasi-particles, i.e. unitary acts of exciting condensed medium, having individual glico-lipo-proteid components combinations, appear on thylakoids surface (nearby chloroplasts internal membranes) of certain photosynthetic organisms. Thylakoids maturation and double layer membrane formation are closely coupled with their components (proteins, lipids, pigments, etc) coming from or controlled by different cellular compartments [73]. Among these compartments are nucleus, cytoplasm and chloroplasts themselves (internal membrane, stroma). Quasi-particles may have individual own partly genetically programmed generalized profile of total function of unitary acts of exciting. That is why, supposedly, interaction of elementary particles (photons, etc) with quasi-particles (glico-lipo-proteid components of certain condensed medium) can be essentially selective. Finally, in the third place, so-called “amino-nucleic conformity language” may be resulting [26], i.e. previously formed and constantly verifiable for amino-acid/codon correspondence to modern UGC-code, but only included in epitope complex (shown on Scheme 1) of 27 - 28 hypothetical retranslosome of chloroplasts (photosynthetic organisms). Let's notice, that in this sense photosynthesis can appear secondary in relation to process of formation of a genetic code. Hereof, it is clear why vIERT-mechanism action, contextually with abovementioned, can be initially associated with “retranslational (photon-/electron/proton-/ion-/…/supramolecular-complex) over-molecular machinery”. Such complex is formed in any photosynthetic organism; however, combination of initiating ERF-components, as well as composition of membrane glico-lipo-protein components, can be individual in each case. At present, we can observe teethed tendency (not devoid of initial stage contradictions) connected with description of possible wave characteristics and properties of biological macromolecules, their parts and complexes. This concerns peptides, proteins, DNA, RNA, DNP-/RNP-complexes and their membrane complexes, membranes, intracellular organelles, ribosomes, etc. Among such characteristics it is possible to find connected both with interaction between of elementary particles (photons of UV-, optical-, IR-, roentgen diapasons and electrons, etc.), quasi-particles (phonon-phonon, phononexitone, etc.) and first with the second (photon-phonon, photon-excitone, etc.) under some conditions [74, 75, 112]. Properties of either particle could begin to come out still before appearance of any present organisms. It concerns early evolutionary stage of autonomous genetic systems formation with maintaining of inter-oriented specific resonant structures (particles of both kinds) and fitting with them fields configurations [26]. Epitopes (and their NEs) sorting, supposedly, proceeds in consequence of interactions mechanisms of self-organizing supramolecular membrane-connected nucleoprotein complexes “retranslosomes”. Such machinery may include nanostructure elements [76] and they are capable to preliminary selective recognition of spatial-geometric configuration, as well as catalysis, transfer and molecular switching. Inside of such dynamic complexes alternating collectivization of electron envelopes of several their elements is possible. Meanwhile, there is periodical formation of structures capable to generate labile assemblies of molecules and lowenergy non-covalent intermolecular bonds. It is possible that electrons 28 - 29 collectivization process is accompanied by downthrow of certain photons combinations and absorption of other combinations (“latent photonic firework”) between molecules and their parts. At least hydrogenic, hydrophobic, ionic, stacking, Van der Waals, dipole-dipole, coordination, donor-acceptor, electrostatic and other bonds participate in this process. Under the hypothesis, epitopes sorting (light dependent) proceeds in hypothetical retranslosomes of photosynthetic organisms. In this regard, both standard and nonstandard (new, modified old) NEs epitopes variants from unique and conservative genome regions are reproduced. VLNS-transfer (with NE inside VLNS), supposedly, is connected with spreading (when exceeding certain threshold level) most strongly reproduced NEs along DNA-containing intracellular organelles (nucleus, mitochondria, chloroplasts). It concerns cells of different organisms in all three biological kingdoms (eukaryotes, prokaryotes, archae-bacteria) of the whole biosphere (including its photo- and non-photosynthetic parts). Such VLNS-transfer between cells of different organisms within community, group of communities’ organisms (involving known and unknown viruses, phages, conditional symbionts, parasites, etc) may be called Genetic Shuttle Feedback system (GSF-system) and concerns the various widely widespread kinds of horizontal transfer of the nucleic information [107]. Abovementioned bunya-, poty- and rhabdoviridae, particularly, belong to such system elements [51]. Nevertheless, NE’s pathway to intron/exon space of newly synthesized transcript or genomes of eukaryote DNA-containing cell organelles may be not fast. Presumably, few/multitude steps are required over evolutionary valuable period for concrete kind of organisms (see below). As a result, nucleotide vectors (with NE inside), probably, may spread not only vertically (from predecessors to descendants) but horizontally (inside GSF-system, between the cells of one or different organisms, including photo- and non-photosynthetic) too. It is considered, that such synchronous pandemic horizontal transfer was especially widespread in early evolution period for extraordinary mutation level. That period is attributed to epoch of generation of collectively metabolizing fore-genetic 29 - 30 systems and unicellulars [77], particularly, purple photosynthetic bacteria [78] when synchronously resistance to horizontal and vertical transfers was formed. Moreover, horizontal transfer is supposed for mitochondrial (and other DNA-containing compartments) introns of mobile I and II types between unrelated species of certain protists as well as protists and cyanobacteria over evolutionary valuable period [79]. Possible connection of hypothetical vIERT/VLNS-trsansfer mechanisms with RNA-editing and other genome expression mechanisms It is supposed that vIERT/VLNS-transfer mechanisms may interact (Scheme 2) with known intracellular genome expression mechanisms. Among those are replication, direct and reverse transcriptions, translation, processing, splicing, different kinds of reparation, RNA-editing, various posttranscriptional and posttranslational mechanisms, etc. Scheme 2 first of all shows possible connection of hypothetical mechanisms with RNA-editing process and formation of different kinds (RNA, DNA, and Protein) polymorphisms. It is known, actually and more potentially, the significant, if not the most part of genome (mRNA, rRNA. tRNA, certain introns, spacers, low molecular RNA, including micro- and small interferential RNA, and repeated regions) are transcribed as well as are edited [26]. It is assumed that high frequency of occurrence of certain nucleotides changes on RNAlevel under editing and back mutations on DNA-level [80] may be interconnected through reverse-transcriptase activity. However, in that case, except of protein polymorphism, as concerning petunia protein rps12 [81], genetic polymorphism should become apparent also. Actually, mutations accumulation level (for example, concerning kinetoplastic СОIII-genes of trypanosome seven species) in widely-edited genes is much higher than in non-edited ones [82]. Eukaryotic evolution, supposedly, is governed by alternative pathways, in which DNA and processing RNA interact permanently [83]. Phenomenon of various kinds’ RNA-editing (mysterious form of processing) is widespread in many eukaryotic organisms and viruses. RNA-editing enzyme was 30 - 31 VLNS-transfer The Genetical Polymorphism RT- and RTlike activity RNA Usual expression Change in: editing: mRNA gRNA U+/__ The Protein Polymorphism due to changes in: NE Hypothetical vIERTmechanism tRNA mRNAs, tRNAs, Other types of RNA editing rRNAs. Checkout functional rRNA significance. gRNA-like (snRNAs/snoRNAs) and oth. Scheme 2 Scheme-2 31 - 32 Possible conjunction of the hypothetical vIERT/VLNS-transfer and the RNA editing mechanisms under the formation of the Protein, RNA and DNA kinds of polymorphisms. NE – one of possible nucleic equivalents of epitope obtained as a result of a hypothetical vIERT-mechanism in one of a DNA-containing cellular structure (organelles). VLNS-transfer – the transmission of a vector-like nucleic sequence. Small RNAs: Trypanosomae kinetoplast (mitochondrial) guiding RNAs (gRNAs) and animal nuclear/nucleolar RNAs (snRNAs and snoRNAs). RT- and RT-like activities – the revertase and similar activities of corresponding cell and viral polymerases (possibly RNA-zymes, DNA-zymes and others) towards short fragments of nucleic sequences. shown for the first time also in prokaryotes, tRNA-specific (tadA) adenosine deaminase E. coli [84]. Mystique is associated with unobviousness of editing sites selective advantages, considerable mechanism uncertainty and unclarity of editing final objectives. It is not clear wherefore cells necessarily permanently maintain and activate high energy-consuming “editing machinery” (particularly in relation of editing parasitic virus mRNA-transcripts). It would be much simple point-wise or over several sites to import nucleotide changes into genes themselves “forever” [52]. It is supposed that editing is a substantial part of biological information transfer process requiring the same accuracy for replication, transcription, translation [33, 85]. Majority of studied RNA-editing forms need informative-matrix component. One of NE variants, not excluded [26], may take part in its formation (see below). Among such components at least the following may turn out to be: 1. gRNAs, easily self-reproducible and self-replicated in minicircular (probable forefathers are plasmids) and maxi-circular DNA components for U-insertiondeletion editing in Trypanosome kinetoplasts. It is interesting, that in related Trypanoplasma borreli gRNAs-genes are positioned at tandem repeats of maxicurcular (200 kb) sequences [86]; 2. introns, in case of А→I (sometimes C→U) editing intron/exon duplex of hairpin dsRNA. Introns are contained in a significant part of tRNA-genes of eukaryotes, bacteria and archaebacterias, promoting their modification and, possibly, descend from auto-splicing introns of I and II groups or exposed expansion of loops of molecules tRNA. In 61 (20%) from 270 known yeast tRNA-genes small introns (14–60 nucleotides) have permanent site. The site is 32 - 33 situated over 1 base from 3’-end of tRNA anticodon stem. Deletion of these introns by special “molecular ruler”-mechanism is associated with recognition by endonucleases of duplex exon/intron bulge-helix-bulge-(B-H-B)-motive with specific secondary structure [42]. Introns pertain to most ancient mobile and speedily evolving genomes regions, are constituent and, vice versa, comprise different genes. This is a great number of protein genes, low molecular RNA genes, and small nuclear/nucleolar RNA included in RNP-complexes (more than 200 species; 104-106 for a cell) of most eukaryotes cellular compartments. Nature of appearance and engagement of snRNAs/snoRNAs-genes in introns, as well as function most snRNPs (including viral) are not known. Such specific snRNPs are involved in synthesize, assembly and maturation of pre-rRNA/pre-mRNA, provide posttranscriptional modifications of those (hundreds of nucleotides are methylated, pseudo-uridilated), rRNA and mRNA export into cytoplasm. Moreover, they control spliceosomes (splicing), ribosomes (proteins translation), and editosomes (RNA-editing). That reminds, however, snRNPsmediated genes expression control [26, 67, 87]. RNA-editing is associated with various effects: appearance and disappearance of sense and stop/start codons; conservatism and hydrophobicity enhancement for protein fragments after transcripts editing; participation in unifying "rewriting" of genes of mitochondria (are more often) and chloroplasts for nucleus [52,88]. Furthermore, RNA-editing is able to: regulate splicing (for example, length of mRNA apolipoprpotein-В transcript: ApoB-100→ApoB-48, etc), shift of reading frame (mRNA, ORF); activate nuclease reactions for virus protection (say for C→U and A→I hyper-editing); provide repair on RNA- and modifications on DNA-levels, etc [26,89]. The reason and the mechanisms of maintaining such a resource-consuming mechanism are not completely clear yet. So, for example, certain site editing observed for some species may be absent in homological transcripts for related species. In this case DNA-site often contains “necessary” nucleotide, and editing is not required [82, 90]. But it is still not known which developments (events) were predecessors of that. 33 - 34 For vIERT-mechanism (Fig. 1, Fig. 2) appearance of oligoribonucleotide NE is most likely. Such NE in composition of retroposon-like VLNS may be integrated but then eliminated or fixed by one/few sites in coding/non-coding (including repeated sequences) part of genome (nucleus, mitochondria, chloroplast). The same, but less likely, is in case of NE deoxyvariant reproduction also. Hypothetically deoxy-NE may develop due to some known or unknown proteins (or nucleozymes) reverse-transcriptase activity. Reverse transcriptase related domains contain certain telomerases, maturases, and nucleotidyltransferases. Homology blocks with seven viral reverse transcriptases were found in mitochondrial genome ascomycetes [41]. RT-activity existence corresponds to the hypothesis of pan-edited mitochondrial cryptogenes (Trypanosome kinetoplasts) substitution for their whole with cDNA-versions retroposing into nucleus. Also it is consistent with the fact of trans-kinetoplastidia of low-copy-number mini-circular DNA into nucleus (mechanism is unknown) under conditions of artificial isolation of cellular structures [86]. Moreover, that is in partial correspondence with the fact of reverse transcription of monomer-length plasmid transcripts (with 3’-ССА-end tRNA-like terminals) of Neurospora spp. fungus containing replicating Mauriceville and Varkud retroplasmids in mitochondria [91]. Both cases of VLNS fixation (with NE ribo- or deoxy-variants) in genome may have not fast consequences and appear then in any form (RNA, DNA, Protein) of cell polymorphisms. Hystohematic barriers of ordinary parenchymal and overbarrier (hematoencephalic, germinative, transplacental) tissues differ strongly. All tissues show at least minimal permeability concerning certain virus, and sometimes, cellular (lymphocyte, etc) agents. Australian authors [92, 93] observed modified germinal configuration (deletions/insertions of individual or several nucleotides) of immunoglobulin genes in gamete genomes of many vertebrates. Based upon these and circumstantial evidence, authors assumed that modifications could be the traces of rearranged V-(D)-J-fragments of immunoglobulin genes integration. Whereas integration itself was a result of transfer retro-copies of such fragments from hyper-mutated lymphocyte and their following 34 - 35 homological recombination with corresponding allelic sequences of gamete haploid genome. Fixation of hyper-mutational phenotype in B-lymphocyte itself was also connected with homological recombination of retro-transcript, but under conditions of intracellular transfer variant. In accordance with our vision, however, VLNStransfer could concern rather shorter nucleic fragment (including NE length) too and be connected not only in concrete B-lymphocyte, but also with low-differentiated precursors of stem hematopoietic cells (LDPhc, see above), further differentiating in various directions including lymphocyte line. Furthermore, site-specific ectopic transfer of mitochondrial yeast II group intron into unrelated RNA by means of reverse splicing is described [67]. That is why, not excluded, ribo-VLNS (with NE inside) can be incorporated into exon-intron space of newly synthesized edited transcripts under trans-splicing-similar process. RNA-RNA-integration under the recombination conditions provides appearance of chimaera’s mitochondrial RNA, particularly 16S RNA of certain animals [94]. Likewise exons 1 and 2 (from different transcripts of psaA gene photosystem I plastids of certain algae) are connected; low-molecular RNA of tscA-gene are involved in this process [36, 95]. Such similarity also concerns “exonization introns” mechanism known in respect of Alu-repeats incorporated into maturating pre-mRNA of certain genes [96]. Various repeated sequences which common share composes major portion of eukaryote genome are interesting in many ways. Alu-repeats, for example, can move into intron/exon space and modify genes functions [96]. Mysterious gRNA-genes responsible for pre-mRNA-editing [86] are localized in tandem-repeated maxicircular kinetoplast sequences of some protozoan (Trypanoplasma borreli, etc). Telomerase RNA-component reverse-transcription provides chromosomes with tandem-repeating telomeric repeats. Spliceosome effectiveness and specificity is often associated with di-, tetra-, penta-, hexa- as well as 74-nucleotidic repeats [87]. In repeated sequences of so-called “junk-DNA” of human origin [97] up to 99% of variable sites (out of their 3 million total number) are detected. 35 - 36 The role of repeated sequences in genome expression is gradually reconsidered. Not excluded, that some of them may turn out to be one of possible sites of NEs primary localization (“depot”). Repeats consensus variants may form from similar but not identical NEs. In turn evolution of many genes, in particular snRNAs/snoRNAs-genes, contained in repeats and introns of protein-coding genes, as well as corresponding snRNPs functions may be dynamically associated with reproduction of repeats consensus variants. Concepts of possible relationship between genetic code evolution, protein’s and nucleic cell components (including introns and repeating sequences) co-evolution and rT-similar mechanism already exist [61]. For integration of NE into transcript we have a case of RNA- or RNA/Protein- polymorphisms. Herewith, fast check for functional value of introduced modifications is possible (Scheme 2). Functionally more significant the modified protein-versions can assure preferential synthesis of respective transcripts. The functional value of modifications is also checked in non-translated RNA (rRNA, tRNA, etc). Probability of retro-transcription for modified whole (large portions) transcripts and their succeeding integration into DNA-genome is not equal to zero, although it is not high under reverse splicing of certain mitochondrial introns in fungi I group. Moreover, subsequent horizontal transfer of introns between cells and organisms is considered possible also [67]. The combination of interactions at RNA-(first) and DNA-(later) levels, in particular, is shown for Zn2+-dependable activation-induced cytidindeaminase (AID) in lymphocytes germinative centers (corresponds to Scheme 2). AID but not Apobec1 cytidindeaminase is active during of SHM formation, gene conversions and CSR in immunoglobulin of activated B-lymphocytes and fibroblasts as well as in some other genes and cells [98]. Under super-expression of these enzymes in E. coli (both are members of RNA-editing cytidindeaminase mammalian family) induced DNAmutations. AID-enzyme is able both to edit RNA and to modify DNA immunoglobulin (DNA-editing model). Transcript is edited in anchoring UGAUCAGUAUA sequence region whereas DNA-modification in mutable GACTAGTAT-nanomer region. There is linkage between these sequences: the part 36 - 37 of nanomer, ACTAGT-hexanucleotide, is complemented to UGAUCA-part of abovementioned (UGAUCAGUAUA) anchoring sequence [26, 99-103]. Modifying effect of AID-enzyme is also supposed with regard to homological IgV-genes of gp120-coding region env-gene hyper-mutative HIV-1 virus [104]. HIV1 hypermutations in chronically infected cell cultures are preferably associated either with C→U modifications under RNA-editing [105] or reverse transcription low accuracy [106] for long-infected cultures. Thus, the hypothetical vIERT/VLNS-transfer mechanisms, probably, can realize in three ways (Scheme 2). The first way is realized only at RNA-level: RNAor RNA- and protein’s polymorphisms are formed. Second way: only at DNA level. Under formation of genes polymorphism NE (consensus variant) pathway in genome may run through its entry firstly into repeated sequences (Alu, oth.), low-molecular RNAs-genes (including miRNAs, siRNAs, oth.) and/or later into intron/exon space of asymmetrically replicated DNA complementary main or lagging strands. In particular, the transcription factors and reciprocal genetic networks of expression with participation miRNAs is regulated [110]; in turn, NE s can take part as in formation miRNAs genes (for the evolutionary period) and expression them, as and in a competition with miRNAs for linkage with 3'-regions of mRNA-targets (by inhibition of translation, RNA degradations). For different genome regions (coding, non-coding), genes (immunoglobulin, “household”, etc) and cells (somatic tissues, germinative, lymphocytes) such pathway and speed of sitespecific NE fixation, obviously, may not coincide. Over evolutionary valuable period, however, combination of both pathways with potential formation of all three polymorphism’ types is more probable (Scheme 2). This requires, presumable, several stages separated by time and site of action for interacting on RNA- and DNA-levels components in different genetic systems. It is unlikely, that these stages coincide for ontogenesis and phylogenesis of individual organism, as well as for more complicated exchanges of genetically valuable material Possible connection of hypothetical mechanisms with some mechanisms of genes/genome expression and biological processes Hypervaria-ty and joint evol cell/vir fragm genomes (norm/pathol; program 37 partition variants) - 38 - Macroevolution? (point Microevolution? Genome Evolution, adaptations [point mutat structural genes, block reorganizations; more likely for intraspecific changes; [vIERT (NE "from below") + VLNStransfer]. Genes Expression Mechanisms (means miRNAs, oth.) mutation regulator genes, large block reorganize-ns genome/chromosomes, horizontal transfer, oth. (ecosystems biosphere: new taxons → new species). (Possible"crossing" ways "from below"/"from above": vIERT + GSF-system (synchronous VLNS-transfer to various biological species). Ecology, the Biodiversity, Food chains. RNA-Editing: use dsRNA-matrix: gRNAs, A→I, C→U. Effects:splicing, rise hydroph/conservatism proteins, shift framework, unification genes, antivir protect, change stop/start/sens codones, oth. Mechanis inter- Hypothetical mechanisms (vIERT/VLNS-transfer, vIERT/GSF-system) genome/intercoding (Nucl, Mt, Chlrpl), retranslation. Phenogenotipical balance. Polymorphism (on Prote- Epigenesis in, RNA, DNA level). Genetical and Epigenetical Non-coding protein exons parts genome (repeats; introns; aspects of pathology. Synchron dynamic maintenance of conservatism in various genomes [vIERT + Horizontal transfer (=GSFsystem)]. Potencial changes in : gRNAs, different small RNAs, repeats, introns, exons, rRNAs, tRNAs, immunoglobulin (BCR, TCR, oth.) molecules families. rRNAs, tRNAs, various smRNAs, gRNAs, oth.) Formation Gen Code/Variety in its frameworks (modern photosynth organisms). New ratio theor RNA-/RNP/DNA-/Protein-Worlds (latent proceeding evolution oligostructures). Cooperating "Languages": Elementary-/Qusielementary-particles, AA/Codone (in structure epitope/NE complex "retranslosome"). Scheme №3. Scheme-3 Possible connection of hypothetical mechanisms with some mechanisms of genes/genome expression and biological processes 38 - 39 in populations and between different organisms of ecological communities. Moreover, all abovementioned concerning vIERT and VLNS-transfer (GSF-system) mechanisms, supposedly, do not exclude their effective (competitively coordinated) of participation in evolution of individual genomes and synchronous processes of micro-/macroevolution in ecological-connected communities [107]. Both processes, however, may be depending from different combinations vertical and specific horizontal methods of transmission and fixation of genetic information. The proposed approach can explain some of the reasons of non-coincidence of different phyletic classifications also. In conclusion we shall result the Scheme 3 reflecting opportunities of perspective analysis for prospective connection of hypothetical mechanisms with group of known/unknown of gene expression processes and mechanisms [111]. Among them there are, at least, following: formation of hypervariability/conservatism in oligonucleotides fragments of genome (including coding/non-coding a parts of genomes) at various biological species; RNA editing; formation of various kinds of polymorphisms (phenogenotipical balance); formations of a genetic code and a variety in its frameworks (including intergenomic/intercoding kinds of retranslation); genetic/epigenetic changes and pathologies; processes of microevolutions of genomes and macroevolutions of organisms. Acknowledgment: Author thank d.b.sc.N.P. Yurina, prof. V. A. Kolb and prof. V.A. Gvozdev for valuable comments and good advice сoncerning article and on the future; c.b.sc. A.A. Vartanian for help in correcting the MS. References: * - In: Gesteland, R.F., Cech, T.R., Atkins, J.F. (Ed.), The RNA World 1999, Cold Spring Harbor Laboratory Press, N.Y. 1. Nashimoto M. The RNA/Protein Symmetry Hypothesis: Experimental Support for Reverse Translation of Primitive Proteins. J Theor Biol 2001, 209: 181-187. 2. Gilbert, W., Souza, S.J. Introns and the RNA World. In: The RNA World* 1999, CSHL- Press, 221-232. 39 - 40 3. Benner S.A., Burgstaller P., Battersby T.R. Did the RNA World Exploit an Expanded Genetic Alphabet? In: The RNA World* 1999, CSHL-Press, 163-183. 4. McKay D.B., Wedekind J.E. Small ribozyme. In: The RNA World* 1999, CSHLPress, 265-286. 5. Feig A.L., Uhlenbeck O.C. The role of metal ions in RNA biochemistry. In: The RNA World* 1999, CSHL-Press, 287-320. 6. Cech T.R., Golden B.R. Building a catalytic active site using only RNA. In: The RNA World* 1999, CSHL-Press, 321-351. 7. Moore P.B. The RNA folding problem. In: The RNA World* 1999, CSHL-Press, 381402. 8. Burkard M.E., Turner D.H., Tinoco I.J. The interactions that shape RNA structure. In: The RNA World* 1999, CSHL-Press, 233-264. 9. Noller H.F., Yusupov M., Yusupova G., Baucom A., Cate J. Translocation of tRNA during protein synthesis. FEBS Letters 2002, 514 (1): 11-16. 10. Altman S., Kirsebom L. Ribonuclease P. In: The RNA World* 1999, CSHL-Press, 351-380. 11. Forterre P. Genomics and early cellular evolution. The origin of the DNA world. C. R. Acad. Sci III 2001, 324 (12): 1067-1076. 12. Gerasimenko L.M., Zhegallo E.A., Zhmur S.I., Rosanov A.Yu., Hoover R. Bacterial paleontology and the studies of the carbonaceous chondrites. Paleontol. J. 1999, 4: 103 – 125 (in Russian). 13. Dyson F. Origins of Life. Cambridge: Cambridge University Press 1985, p.125-127. 14. Di Giulio M., Medugno M. Physicochemical optimization in the genetic code origin as the number of codified amino acids increases. J. Mol. Evol. 1999, 49 (1): 1-10. 15. Issac R, Ham Y.W, Chmielewski J. The design of self-replicating helical peptides. Curr. Opin. Struct. Biol. 2001, 11 (4): 458-63. 16. Lee D.H., Granja J.R, Martinez J.A., Severin K., Ghadri M.R. A self-replicating peptide. Nature 1996, 382 (6591): 525-528. 17. Rode B.M. Peptides and the origin of life. Peptides 1999, 20 (6): 773-86. 18. Bartel D.P. Re-creating an RNA Replicase. In: The RNA World* 1999, CSHL-Press, 143-163. 19. Li Y., Breaker R.R. Deoxyribozymes: new players in the ancient game biocatalysts. Curr. Opin. Struct. Biol. 1999, 9 (3): 315-323. 20. Breaker RR. Natural and engineered nucleic acids as tools to explore biology. Nature 2004, 432 (7019): 838-45. 21. Butcher SE, Burke JM. A photo-cross-linkable tertiary structure motif found in functionally distinct RNA molecules is essential for catalytic function of the hairpin ribozyme. Biochemistry 1994, 33 (4): 992-993. 22. Liu Y., Sen D. Light-regulated catalysis by an RNA-cleaving deoxyribozyme. J. Mol. Biol. 2004, 341 (4): 887-892. 23. Deichman A.M. One of the variants of point mutations is possibly triggered by the per-epitope reverse translation. The hypothetical concept. MS, VINITI 1993, Depon. №1502-B93, 56p. (in Russian). 24. Deichman A.M. The black box of genetic code. Chemistry and Life J 1994, 11: 28 – 33 (in Russian). 25. Deichman A.M., Smirnov A. Yu. New hypothetical mechanisms and the multi-level adaptation of cells. 3rd Internat. Congr. weak and super-weak emissions: selected works. St-Petersburg 2003, 79 – 83 (In Russian). 26. Deichman A.M., Choi W.C., Baryshnikov A.Yu. RNA editing. Hypothetical mechanisms. Publishing-House “Practical medicine” (www.medprint.ru), Moscow 2005, (in English 265p, and in Russian 302p.). 27. Altshtein A.D. The origin of genetic system: a hypothesis of progenies. Mol. Biol. (Moscow) 1987, 21 (5): 1411-1429 (in Russian). 40 - 41 28. Altshtein A.D., Efimov A.V. Physicochemical basis for the origin of the genetic code: stereo-chemical analysis of the amino acid and nucleotide interaction based on the progeny hypothesis. Mol. Biol. 1988 (Moscow), 22 (5): 1411- 1428 (in Russian). 29. Nelsestuen G.L. Amino Acid-Directed Nucleic Acid Synthesis. J. Mol. Evol. 1978, 11: 109-120. 30. Woese C.R. Codon recognition: the allosteric ribosome hypothesis. J. Theor. Biol. 1970, 26 (1): 83-88. 31. Crick F.N., Orgel L.E. Directed panspermia. Icarus1973, 19: 341-346. 32. Burge C.B., Tuschl T., Sharp P.A. Splicing of precursors to mRNA by spliceosomes. In: The RNA World* 1999, CSHL-Press, 525-560. 33. Deichman A.M. RNA editing. Publishing-House “Rusaki” 2001 (Moscow), 131p (in Russian). 34. Schabalkin I.P., Schabalkin P.I., Yaigubov A.S. Evolution of genetically alphabet and amino acid code. Journal of a developmental biochemistry and physiologies 2002, 39 (5): 488-494 (in Russian) 35. Di Giulio M. The non-monophyletic origin of the tRNA molecule. J. Theor. Biol. 1999, 197 (3): 403-414. 36. Odintsova M.S., Yurina N.P. Genome plastids of highest plants and algae(s): structure and function. Mol. Biol. 2003, 37 (5): 1-16 (in Russian) 37. Maizels N., Weiner A.M. The genomic Tag Hypothesis: What molecular fossils tell us about of evolution of tRNA? In: The RNA World* 1999 CSHL-Press, 79-81. 38. Lim V., Venclovas C., Brimacombe R., Mitchell P., Muller F. How are tRNA arranged in the ribosome? An attempt to correlate the stereochemistry of the tRNAmRNA interaction with imposed by the ribosomal topography. Nucl. Acid. Res. 1992, 20 (11): 2627-2637. 39. Steitz T.A. RNA recognition by proteins. In: The RNA World* 1999, Cold Spring Harbor Laboratory Press, 427-450. 40. Zaenger W. Principles of Nucleic Acid Structure. Publishing-House Springer-Verlag, New-York-Berlin-Heidelberg-Tokyo 1984, 584p. 41. Kuzmin E.V., Zaitseva G.N. Mitochondrial genome organization and expression. In: Nesterov P.V. (Ed.), the Totals Sciences and Techniques, Publishing-House VINITI (Moscow) 1987, vol. 6:.15-55 (in Russian). 42. Trotta C.R., Abelson J. tRNA-splicing: an RNA add-on or an ancient reaction? In: The RNA World* 1999, CSHL-Press, 585-608. 43. Tchaikovsky Yu.V. Evolution. Publishing-House “Systemic Examinations Center” (Institute of Natural History and Technique RAS, Moscow), 2003, 472p. (in Russian). 44. Reichert A.S, Morl M. Repair of tRNAs in metazoan mitochondria. Nucleic Acids Res. 2000, 28 (10): 2043-2048. 45. Spahn C.M, Remme J., Schafer M.A., Nierhaus K.H. Mutational analysis of two highly conserved UGG sequences of 23S rRNA from Escherichia coli. J. Biol. Chem. 1996, 271 (51): 32849-32856. 46. Philippovich I.I., Nozdrina V.N., Svetelukin V.V., Oparin A.P. The study of localization of the transcription and translation systems in the fine structure of chloroplasts in relation to the grana formation. In: Neufax S.A., Troshin A.S. (Eds.), Molecular genetics of mitochondria. Publishing-House “Nauka” (Leningrad) 1977, 11 – 20 (in Russian). 47. Puglisi J.D., Williamson J.R. RNA interaction with small ligands and peptides. In: The RNA World* 1999, CSHL-Press, 403-426. 48. Herbert A., Wagner S., Nickerson J.A. Induction of protein translation by ADAR1 within living cell nuclei is not dependent on RNA editing. Mol. Cell. 2002, 10 (5): 12351246 41 - 42 49. Simpson L., Maslov D.A., Blum B. RNA editing in Liechmania mitochondria. In: Benne, R. (Ed.), RNA editing. The alteration of protein coding sequences of RNA. Ellis Harwood Ltd. (London) 1993 53-85. 50. Frolov V.A., Puxlianko V.P. Morphology of mitochondria cardiomycetes in norm and pathology. University of Friendship of the Peoples (Moscow) 1989, 142р. (in Russian). 51. Bishop D., Emerson S.U. Rhabdoviridae and Bunyaviridae. In: Fields, B.N., Knipe, D.M. (Eds.), Virology, Publishing-House “MIR” (Moscow) 1989, vol.2: 366-445 (in Russian). 52. Odintsova M.S., Yurina N.P. RNA editing in chloroplasts and mitochondria of plants. Plants Physiology 2000, 47: 307 – 320 (in Russian). 53. Alfonzo, J.D., Blanc, V., Estevez, A.M., Rubic, M.A., Simpson, L. C to U editing of the anticodon of imported mitochondrial tRNA (Trp) allow decoding of the UGA stop codon in Leichmania tarentolae. EMBO J. 1999, 18 (24): 7056-7062. 54. Korochkin L.I. The biology of individual development. Publishing-House of the Moscow University 2002, 263p. (in Russian) 55. Zorov D.B. Mitochondrial transport of nucleic acids. Participation of the benzodiazepine receptor. Biochemistry (Moscow) 1996, 61 (7): 1320 – 1332. (in Russian). 56. Zorova L.D., Krasnikov B.F., Kuzminova A.E., Polyakova I.A., Dobrov E.N., Zorov D.B. Virus-induced permeability transition in mitochondria. FEBS Letters 2000, 466: 305-309. 57. Khaitov R.M., Ignatieva G.A., Sidorovich I.G. Immunology. Publishing-House “Medicine” 2000, 1-432 (in Russian). 58. Deichman A.M. AIDS and Hyper-variability: Hypothetical Mechanisms. In: Norrby, E., Brown, F., Chanok, R.M., Ginsberg, H.S. (Eds.) 1994, Vaccina-94, CSHLPress, N.Y., 291-293. 59. Deichman A.M. One Variant of Point Mutation is Possible Triggered by Reverse Translation of Individual Epitope: A Hypothetical Concept. Hematology Reviews: Soviet Medical Review (Sect. C) 1997, Harwood Acad. Press, Gordon and Breach, 7 (3): 57-79. 60. Burnet F. M. The Integrity of the body. Publishing-House “Harvard University Press”, Cambridge 1962, 180p. 61. Biro J.C. Seven fundamental, unsolved questions in molecular biology. Cooperative storage and bi-directional transfer of biological information by nucleic acids and proteins: an alternative to "central dogma". Med Hypotheses 2004, 63 (6): 951-962. 62. Cook N.D. The case for reverse translation. J. Theor. Biol. 1977, 64: 113-135. 63. Craig R. The theoretical possibility of reverse translation of proteins into genes. J. Theor. Biol. 1981, 88: 757-760. 64. Mekler L.B. Mechanism of biological memory. Nature 1967, 215 (100): 481-484. 65. Mekler L.B. The origin of living cells: the evolution of biologically meaning molecules as a reflection of the transition from chemical to biologic evolution. J. All-Union Chemistry Society by Mendeleyev 1980, D.I., 25: 462 (in Russian). 66. Odintsova M.S., Yurina N.P. Genomics and evolution of the cell’s organelles. Genetics (Moscow) 2005, 41 (9): 1-12 (in Russian). 67. Lambowitz A.M., Caprara M.G., Zimmerly S., Perlman P.S. Group I and group II ribozyme as RNPs: clues to the past guides to the future. In: The RNA World* 1999, CSHL-Press, 451-486. 68. Yurina N.P., Odintsova M.S. The comparative characteristics of structural organization of the chloroplast genomes and plant mitochondria. Genetics (Moscow) 1998, 34 (1): 1383 – 1389 (in Russian). 69. Deichman A.M. Possible formation of amino-nucleic correspondence (epitope) in chloroplasts of photosynthesizing organisms under the influence of different factors of the biosphere. 2nd Congress of Russian Biophysicists 1999, 3: 778 – 779 (in Russian). 42 - 43 70. Lima-de-Faria A. Evolution without selection. Form and function by autoevolution. Moscow 1991, Publishing-House "World", (in Russian). 71. Haken H. The quantum theory of solid body. Publishing-House “Science” (Moscow) 1980, 319-396 (in Russian). 72. Kizel V.A. Physical causes of asymmetry in living systems. Publishing-House “Science” (Moscow) 1985, 52-55 (in Russian). 73. Wettstein D. Discovery of a protein required for photosynthetic membrane assembly. PNAS 2001, 98 (7): 3633-3635. 74. Chirkova E.N. Immune-specificity of the wave information. Publishing-House “New Center” (Moscow) 1999, 303p. (In Russian). 75. Semenov S.N. Molecular-mechanical model of functioning and structure of biological membranes. Introduction in quantum phonons biology of membranes. – SciTecLibrary.ru – 2007. – (http://www.sciteclibrary.ru/rus/catalog/ pages/6013.html) (in Russian). 76. Zorkiy P.M., Lubnina E. Supramolecular chemistry: origin, development, perspectives. Publish.-House “Herald of Moscow’s University” (part 2) 1999, 40 (5): 45-46 (in Russian). 77. Woese C.R. Interpreting the universal phylogenetic tree. PNAS 2000, 97 (15): 83928396. 78. Woese C.R., Guptar R. Are archae-bacteria merely derived “prokaryotes”? Nature 1981, 289 (5793): 95-96. 79. Odintsova M.S., Yurina N.P. Mitochondrial genome of protists. Genetics (Moscow) 2002, 38 (6): 773-778. (In Russian). 80. Landweber L.F, Gilbert W. RNA-editing as a source of genetic variation. Nature 1993, 363: 179-182. 81. Lu B., Wilson R.K., Phreaner C.G., Mulligan R.M., Hanson X. Protein polymorphism generated by differential RNA editing of plant mitochondrial rps12 gene. Mol. Biol. Cell. 1996, 16 (4): 1543-1549. 82. Grienenberger J.M. RNA-editing in plant organelles. In: Benne R. (Ed.) RNAediting: The alteration of protein coding sequences of RNA, Ser. Mol. Biol., Ellis Harwood Ltd. (London) 1993, 154-180. 83. Herbert A., Rich A. RNA processing and the evolution of eukaryotes. Nat. Genet. 1999, 21 (3): 265-269. 84. Wolf J., Gerber A.P., Keller W. TadA, essential tRNA-specific adenosine-deaminase from E.coli. EMBO J. 2002, 21 (14): 3841-3851. 85. Jakubowski, H. (1995). Proofreading in vivo. Editing of homocystein by amynoacyltRNA synthetase in Escherichia Coli. J. Biol. Chem., 270 (30): 17672-17673. 86. Simpson L. RNA editing – an evolutionary perspective. In: The RNA World* 1999, CSHL-Press, 585-608. 87. Yu Y.T., Scharl E.S., Smith C.M., Steitz J.A. The growing world of small nuclear ribo-nucleoproteins. In: The RNA World*1999, CSHL-Press, 487-524. 88. Stuart, K. (1998). RNA-editing: trypanosome rewrites the genetic code. Verh. K. Acad. Geneeskd. Belg., 60 (1): 63-74. 89. Yang Y., Ballatori N., Smith H.C. ApoB mRNA editing and the reduction in synthesis and secretion of the atherogenic risk factor, apoB-100 can be effectively targeted through TAT-mediated transduction. Mol. Pharmacol. 2002, 61 (2): 269-276. 90. Tsudzuki, T., Wakasugi, T., Sugiura, M. Comparative analysis of RNA editing sites in higher plant chloroplast. J. Mol. Evolve, 2001, 53 (4-5): 327-332. 91. Kennell J.C., Wang H., Lambowitz A.M. The Mauriceville plasmid of Neurospora spp. uses novel mechanisms for initiating reverse transcription in vivo. Mol. Cell. Biol. 1994, 14 (5): 3094-3107. 43 - 44 92. Blanden R.V., Rothenfluh H.S., Zylstra P., Weiller J.F., Steele E.J. Signature of somatic hypermutations appears to be written into the germinal IgV segment repertoire. Immunol. Rev. 1998, 162: 117-132. 93. Steele E.J., Lindly R.A., Blanden R.V. Lamarck’s Signature. How retrogenes are changing Darwin’s natural selection paradigm. Publishing-House “Allen and Unwin” (Canberra), Series frontiers of science (Paul Davies Ed.) 1998, 231. 94. Villegas J., Muller I., Arredondo J., Pinto R., Burzio L.O. A putative RNA editing from U to C in a mouse mitochondrial transcript. Nucl. Acid. Res. 2002, 30 (9): 1895901 95. Odintsova M.S., Yurina N.P. Chloroplast genomic of land plant and algae. In: Giardi, M.T., Piletska, E.V. (Eds.), Biotechnological Applications of Photosynthetic Proteins, Georgetown, Texas, USA, 2006, 72-88. 96. Dagan T., Sorek R., Sharon E., Ast G., Graur D. Alu-Gene: a database of Aluelements incorporated within protein-coding genes, Oxford University Press 2004, 489492. [Nucleic Acids Res., (Database issue): D489–D492 DOI: 10.1093/nar/gkh132]. 97. Tarantul V.Z. The human genome. Publish.-House “Tongues of slavic culture” (Moscow) 2003, 390p. (in Russian). 98. Eto T., Kinoshita K., Yoshikawa K., Muramatsu M., Honjo T. RNA-editing cytidine deaminase Apobec-1 is unable to induce somatic hypermutations in mammalian cells. PNAS 2003, 100 (22): 12895-12898. 99. Foster S.J., Dorner T., Lipsky P.E. Somatic hypermutations of the VkJk rearrangements: targeting of RGYW motifs on both DNA strands and preferential selection of mutated codons within RGYW motifs. Eur. J. Immun. 1999, 29 (12): 40114021 100. Jakobs, H., Bross, L. Towards an understanding of somatic hypermutations. Curr. Opin. 2001, 13 (2): 208-218. 101. Kinochita K., Honjo T. Linking class-switch recombination with somatic hypermutations. Nat. Rev. Mol. Cell. Biol. 2001, 2 (7): 493-503. 102. Liu M.M., Zhu M., Schartt M.D. Sequence dependent hypermutations of immunoglobulin heavy chain in cultured B-cells. PNAS 1997, 94 (10): 5284-5289. 103. Muramatsu, M., Sankaranand V.S., Anant S. et al. Specific expression of activationinduced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in GCs B-cell. J. Biol. Chem. 1999, 274 (26): 18470-18476. 104. Suslov, K.V. Does AID aid AIDS? Immunol. Lett. 2004, 91 (1), pp.1-2. 105. Bourara K., Litvak S., Araya A. Generation of G-to-A and C-to-U changes in HIV-1 transcripts by RNA editing. Science 2000, 289 (5484): 1564-6. 106. Berkhout B., Das A.T., Beerens N. HIV-1 RNA editing, hypermutations, and errorprone reverse transcription. Science 2001, 292 (5514): 7. 107. Nazarov V.I. Non-Darwin's evolution: Change of evolution model. Publishing-House “ComBook” (Moscow) 2005, 520p, (in Russian). 108. Joyce G.F., Orgel L.E. Prospects for understanding the origin of the RNA World. In: The RNA World 1999, CSHL-Press, 49-79. 109. Yarilin A.A. The basics of immunology. Publishing-House “Medicine” (Moscow) 1999, 608p. (In Russian). 110. Shivdasani R.A. MicroRNAs: regulators of gene expression and cell differentiation. Blood 2006, 108 (12): 3646-53. 111. Deichman A.M. Hypothetical root of genome metabolism mechanisms. St.Petersburg: the Intellectual Forum «Open Door », Federal Medical and Biologic Agency Fed-Stat-Unit-Enterprise of Sci-Res-Inst «Industry and Marine Medicine", 2007, pp. 260-268. (in Russian) 112. Kolman E.V. Laser stimulation of biological objects as process of interaction of nonequilibrium open systems. Physics in biology and medicine // Yekaterinburg: Collection of works of the Second Russian Conference «Physics in biology and 44 - 45 medicine» ( http://virlib.eunnet.net/conferences/phbm/index.html ). – 2001 г. (in Russian) 113. Fedorov V.I. Principles of the organization and functioning of alive systems. P.2. Managing systems of an organism. Novosibirsk: Publishing house NGTU, 2003, 142с. (In Russian). 114. Savinov A.B. Biosystemology: system bases of the theory of evolution and ecology. Nizhniy Novgorod: Publishing house of the Nizhniy Novgorod state university, 2006, 205с. (In Russian) 45