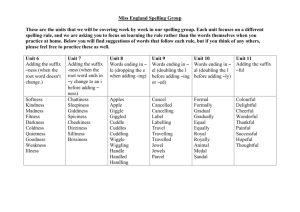
Sequence Query Processing
advertisement

Sequence Query Processing
To processing sequence queries, we need to first based on the sequence of features in the images
and construct an efficient indexing system. Next, we shall search for similar sequences and
compare to the target query sequence and rank them according to a certain measure. In the
following we shall discuss the Index construction procedure and sequence query processing
Technique and follow by an demonstration.
Index Construction Procedure
The index construction procedure consists of the three steps:
1. normalization
2. categorization
3. suffix tree construction
The short description for the normalization step is shown in
in (norm.ps).
At the categorization step, we convert each element value into
the symbol of the corresponding leaf node of TAH.
The detailed description of this step can be found in Appendix A.
At the index construction step, we extract every possible suffix
from each sequence and insert it into a suffix tree. The concept
and the structure of a suffix tree is shown in Appendix A.
Sequence query processing
The sequence query processing procedure consists of the following
three steps:
1. normalization
2. suffix tree traversal
3. post-processing
At the normalization step, we convert each element value of a
query sequence into its normalized value. For this step, we maintain
the average and the standard deviation values of each dimension.
At the suffix tree traversal step, we traverse the suffix tree
to find a set of candidate subsequences using lower-bound time warping
distance function. The concept, the definition, and the lower-bound
definition of the time warping distance are shown in Appendix A.
The false-hit is detected and discarded at the post-processing step
where actual data sub-sequences corresponding to candidate subsequences
are retrieved from the database and compared with a query sequence.
The method for handling the nearest-neighbor query is described
in (nn.ps)
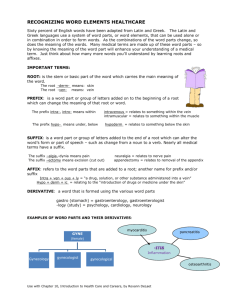
Appendix A
Searches for patients with similar temporal characteristics can augment
the process of patient care by providing physicians with insight into
the treatment of previous patients with similar medical conditions. For
example, an oncologist might search the database for patients with
similar tumor evolution patterns to find the optimal course of
treatment.
Supporting similarity searches of temporal sequences requires the
definition of a similarity measure. Unfortunately, defining such a
similarity measure for medical data sequences is difficult because
sequences that are qualitatively the same may be quantitatively
different; the reasons are twofold: 1) the compared sequences may be of
different lengths, making it difficult to embed sequences in a metric
space and use a Euclidean distance measure; and 2) the sampling rates
of compared sequences may be different, negating the utility of
similarity measures like cross-correlation. In the area of speech
recognition, these problems have been approached using time warping,
allowing sequences to be stretched or compressed along the temporal
axis [RJ93]. Analogous methods can be used to measure the similarities
between sequences of medical data; but without an appropriate indexing
scheme, query processing performance suffers as the total number and
length of sequences increase.
Spatial access methods, such as R-trees, have been used as index
structures for the fast retrieval of similar sequences. While spatial
methods work well using Euclidean distances as a similarity measure,
these methods may produce false dismissals when using time warping
distance as a metric. A false dismissal refers to a sequence that is
actually similar to the query sequence but which is judged by the
system to be dissimilar [AFS93]. Indexing methods predicated upon the
triangular inequality (TI) may generate false dismissals when a
distance function not satisfying the TI is used as a similarity measure
[YJF98]. As the time warping distance metric does not satisfy the TI,
spatial access methods using this metric are subject to false
dismissals.
Suffix trees have been used extensively as an index structure to
find sub-strings matched exactly to a given query string [Ste94], and
are extremely useful for subsequence matching as all possible
subsequences in a sequence are easily determined. Unlike spatial access
methods, suffix trees do not assume a specific geometry, nor a distance
function. As such, suffix trees are a suitable candidate for an
indexing structure based on time warping distances. However, for a
suffix tree to be used in such a manner, the following problems need to
be solved: 1) a suffix tree is built on data sequences whose elements
take values from finite alphabets – but most elements of medical data
sequences have continuous real values; and 2) conventional suffix trees
are used for finding exactly matched sub-strings, so suffix tree
algorithms need to be adapted to search for similarly matched
subsequences.
We propose an indexing technique for similarity searching of
multi-dimensional data sequences. In our approach, similarities between
sequences are measured by the time warping distance. To achieve fast
retrieval of similar sub-sequences without false dismissals, a diskbased suffix tree is used as an index structure. In addition, we
introduce a categorization technique to compress the suffix tree,
making the index structure efficient and scalable.
A.1 Related work
Several approaches for the fast retrieval of similar sequences have
recently been proposed, many of them focusing on one-dimensional data
sequences and using Euclidean distance as a similarity measure. In
[AFS93], sequences of single-dimensional real numbers are converted
into the frequency domain by a discrete Fourier Transform (DFT) and are
subsequently mapped into multi-dimensional points that are managed by
an R*-tree; the technique is extended to locate similar subsequences in
[FRM94]. As this approach uses Euclidean distance as the similarity
measure metric, data sequences of different lengths or different
sampling rates cannot be matched.
The access methods of [BYO97, YO96, YJF98] permit the matching of
sequences of different lengths. [BYO97] presents a modified version of
an edit distance, considering two sequences as being matched if a
majority of the elements match; this approach is extended to the
matching of multi-dimensional data sequences in [YO96]. In [YJF98], the
time warping distance is used as a similarity measure with a two-step
filtering process: a FastMap index filter followed by a lower-bound
distance filter [FL95]. These different approaches focus on whole
sequence searches and use index structures based on the triangular
inequality, therefore leading to possible false dismissals.
Similarity-based temporal sequence searching has also been
addressed in the area of video databases. Work in [SYV97, LSYR98]
describes similarity subsequence searching of video clips; however,
this research focuses primarily on multimedia modeling, and applies
spatial indexing techniques with sequential scanning to perform
matching.
A.2 Similarity measure
In our approach, similarities between sequences are measured by the
time warping distance metric [RJ93]. Time warping is a generalization
method for comparing discrete sequences to sequences of continuous
values, and has been used in the matching of voice, audio, and medical
data (e. g., electrocardiograms). To find the minimum difference
between two sequences, time warping is used to map each element of a
sequence to one or more neighboring elements of a secondary sequence.
Let X = <x1,…,xn> be a multi-dimensional temporal data sequence and let
x = <x1,…,xm> be a feature vector consisting of m features. Then for two
multi-dimensional sequences, X and Y, the multi-dimensional time
warping distance Dmtw(X,Y) is defined as:
Dmtw(X,Y) = Dmbase(X[1], Y[1])
+ min {Dmtw(X,Y[2:-]), Dmtw(X[2:-],Y), Dmtw(X[2:-],Y[2:-])}
Dmbase(x,y) = i=1..m wi * |x[i]-y[i]|
where m is the number of features and wi is the weight of the ith
feature, where (w1 + … + wm = 1).
In the definition of Dmtw(X,Y), X[p] represents the pth element of
X, and X[p:r] denotes the subsequence of X including elements in
positions p through r. The notation X[p:-] is used for the suffix of X
starting at the pth element. That is, X[p:-] is identical to X[p:|X|]
where |X| is the number of elements contained in X. Dmtw(X,Y) can be
efficiently calculated with algorithmic complexity O(m|X||Y|) using a
dynamic programming technique based on recurrence relations [BC96].
A.3 Index construction
We propose the use of a suffix tree as an index structure for
similarity searches based on a time warping distance function. Unlike
spatial access methods, a suffix tree does not assume any geometries
nor an underlying distance function, and therefore, can guarantee the
absence of false dismissals if actual distances are always lowerbounded within the indexing space. We first present the definition and
structure of a suffix tree.
The suffix tree structure is based upon tries and suffix tries. A
trie is an indexing structure used for indexing sets of keywords of
varying sizes. A suffix trie is a trie whose set of keywords comprises
the suffixes of sequences. Nodes of a suffix trie with a single
outgoing edge can be collapsed, yielding a suffix tree [Ste94]. Each
suffix of a sequence is represented by a leaf node. The concatenation
of the edge labels on the path from the root of the tree to the
internal node ni represents the longest common prefix of the suffixes
represented by the leaf nodes under ni . We denote the labels on the
path connecting nodes ni and nj as label(ni, nj).
Based on these definitions and notation, we now present the index
construction methodology, which consists of three steps: preprocessing, categorization, and suffix tree construction.
Pre-processing: In this step, the raw data sequences are converted to
sequences of feature vectors. To obtain a feature vector for an image,
the automatic knowledge-based image segmentation is applied; the
resultant contours are then used to generate a set of spatial features.
As an example, consider the lung image sequence containing lung
lesions, shown in Figure 1. To characterize the lesion evolution
pattern, each lung lesion is represented by three features: area,
perimeter, and circularity. After pre-processing, a sequence of three
feature vectors, X = <(2.1, 3.0, 0.3), (3.9, 5.3; 0.4), (8.1, 9.3,
0.9)>, is obtained to represent the image sequence.
Figure 1: Visual stream representation for a temporal CT lung image sequence, where the size of the tumor is increasing over time.
Three snapshots of a tumor's progression are depicted; for each image, appropriate features are extracted from the segmented lesion,
including the area, perimeter, and circularity.
Categorization: In this step, every feature vector is converted to a
corresponding symbol using the categorization (i.e., clustering)
technique of MDISC [CC94, CCHY96]. MDISC maps a large set of element
values into a much smaller set of categories, and has the following
benefits: 1) the algorithm considers both value and frequency
distributions, thus generating more accurate categories than equallength interval categorization methods; and 2) MDISC is easier to
implement than maximum entropy categorization methods [CC94]. Based on
MDISC, each category is assigned a unique symbol, denoted as C = ([lb1,
ub1],…,[lbm, ubm]), where lbi and ubi are the lower and upper bounds of
categories for the ith feature. Thus, for any element included in a
category, C, the ith feature of the element is contained between lbi and
ubi. For example, given two categories
C1 =( [lb1=1.0, ub1=4.0], [lb2=1.0, ub2=6.0], [lb3=0.1, ub3=0.5]) and
C2 =( [4.1,10.0], [6.1,12.0], [0.6,1.0]), the sequence X for Figure 1
can be converted to Xc = <C1, C1, C2>. Similarly, a second sequence, Y =
<(6.5, 9.2, 0.8), (3.5, 5.8, 0.4)>, with the corresponding
categorization sequence being Yc = <C2, C1>. The sequences of feature
vectors are converted to sequences of finite symbols after
categorization, greatly reducing the overall size of the suffix tree
[PCYH00].
Suffix tree construction: Based on the sequences of symbols obtained
from the previous step, a suffix tree is constructed.
To handle large set of sequences, we employ an incremental disk-based
suffix tree construction method, whereby the tree is constructed by
performing a series of binary merges of suffix trees of increasing size
[PRC94]. Two suffix trees for X and Y are merged with complexity O(|X|
+ |Y|); hence the suffix tree for M sequences is constructed with
complexity O(M L) where L is the average length of M sequences. The
total number of nodes in a suffix tree is constrained due to two
factors: there are O(M L) leaf nodes, and the degree of any internal
node is at least two. Therefore, the maximum number of nodes and
overall space requirement of the suffix tree is linear in M L [Ste94].
As an example, consider the two categorized sequences Xc and Yc. Figure
2 shows the suffix tree constructed from these two categorized
sequences.
Figure 2: Suffix tree from two categorized sequences Xc = <C1, C1, C2> and Yc = <C2, C1>.
The three suffixes X[1:-] = <C1, C1, C2>, X[2:-] = <C1, C2>, and X[3:-] = <C2> from Xc are
represented by the three leaf nodes n3, n4, and n6, respectively. Likewise, the two
suffixes Y[1:-] = <C2, C1> and Y[2:-] = <C1> from Yc are represented by the two leaf nodes
n7 and n2. $ denotes the end marker of a suffix.
A.4 Query processing
Given a multi-dimensional query sequence, Q, the goal of query
processing is to find the nearest k similar subsequences from a set of
M multi-dimensional medical data sequences, {X1,…,XM} where Xi is of
arbitrary length. The query processing consists of three steps (Figure
3): pre-processing, index searching, and post-processing.
Figure 3: Query processing for similarity-matching of temporal data sequences.
Pre-processing: Based on the given feature (i.e., query attribute)
names, their associated weights, and the type of query sequence, the
query is converted into a sequence of feature vectors. For example,
Figure 4 shows the query for a lung image sequence with specific
feature names (area, perimeter, circularity), their associated weights
(area=0.6, perimeter=0.2, circularity=0.2), and the desired number of
answers (k = 2). From pre-processing, a query sequence with two feature
vectors, Q= <(3.0, 4.2, 0.4), (8.0, 8.5, 0.8)>, is obtained.
Figure 4: Visual query for finding patients having similar lung tumor evolution patterns.
The lesions are segmented and three features (area, perimeter, circularity) are
calculated. Weights for features and the desired number of retrieved similarity-matched
answers are specified in this query.
Index searching: Recall that the categorization process partitions the
sequence elements into groups. The edges of the suffix tree are labeled
by the groups of alphabetic symbols. The range of the group is
represented by lower-and upper-bound values. Based on the time warping
distance function, Dmtw(), we define Dmtw-lb() based on the lower-bound
values and Dmtw-ub() based on the upper-bound values. For two multidimensional sequences, X and Y, Dmtw-lb(X, Y) Dmtw(X, Y) Dmtw-ub(X, Y) is
always satisfied.
To find subsequences that may be included in the set of k-nearest
subsequences, depth-first traversal is performed on the suffix tree.
Let n0 represent the root of the suffix tree, and UBk the kth largest
upper-bound distance in the set of candidate answers found thus far
(initialized to infinity). Starting at n0, each child node, ni, is
inspected; if Dmtw-lb(label(n0, ni), Q) UBk, the search algorithm adds
label(n0, ni) to the set of candidate answers. As these new candidate
answers are added, the value of UBk is updated accordingly. To reduce
the search space, a branch pruning technique is applied to node ni to
determine if further traversal is required [PCYH00]. This index
searching step produces a set of k’( k) candidate answers.
Consider the query sequence, ~ Q, shown in Figure 4, and the set
of data sequences represented by the suffix tree in Figure 2. The
search algorithm starts by inspecting node n1. As Dmtw-lb(label(n0, n1), Q)
UBk (= ), then label(n0, n1) is inserted into the set of candidate
answers. Two more subsequences, label(n0, n3) and label(n0, n4), are
added to the set of candidates during the subsequent stages of the
search. The final set of candidates resulting from the traversal of the
complete suffix tree is {label(n0, n1), label(n0, n3), label(n0, n4)}.
Post-processing: For each candidate found during the index searching
step, the corresponding subsequences are retrieved and the exact
distances are computed using the distance function Dmtw(). Subsequences
are ranked according to these distances and the nearest k subsequences
are returned as the final answers. For example, given the set of
candidates obtained from index searching, the three subsequences
Y[2:2], X[1:1], and X[2:2], are retrieved from label(n0, n1), and their
distances to Q are calculated. Similarly, the two subsequences, X[1:3]
and X[2:3], are determined from label(n0, n3) and label(n0, n4),
respectively. Based on the distance values to Q, the nearest two
subsequences, X[2:3] and X[1:3], are returned as the final answers.
References
[AFS93] R. Agrawal, C. Faloutsos, and A. Swami. Efficient similarity
search in sequence databases. In Proceedings FODO, 1993.
[BC96] D. J. Berndt and J. Clifford. Finding patterns in time series: A
dynamic programming approach. In Advances in Knowledge Discovery and
Data Mining, 1996.
[BYO97] T. Bozkaya, N. Yazdani, and M. Ozsoyouglu. Matching and
indexing sequences of different lengths. In Proceedings of the ACM
CIKM, 1997.
[CC94] W. W. Chu and K. Chiang. Abstraction of high level concepts from
numerical values in databases. In Proceedings of the AAAI Workshop on
Knowledge Discovery in Databases, 1994.
[CCHY96] Wesley W. Chu, Kuorong Chiang, Chih-Cheng Hsu, and Henrick
Yau. An error-based conceptual clustering method for providing
approximate query answers. In Communications of ACM, Virtual Extension
Ed., 1996.
[FL95] C. Faloutsos and K. Lin. FastMap: A fast algorithm for indexing,
data-mining and visualization of traditional and multimedia datasets.
In Proceedings of the ACM SIGMOD, 1995.
[FRM94] C. Faloutsos, M. Ranganathan, and Y. Manolopoulos. Fast
subsequence matching in time-series data-bases. In Proceedings of the
ACM SIGMOD, 1994.
[LSYR98] K. L. Liu, P. Sistla, C. Yu, and N. Rishe. Query processing in
a video retrieval system. In Proceedings of the 14th International
Conference on Data Engineering, Orlando, Florida, USA, February 1998.
IEEE, IEEE Computer Society Press.
[PCYH00] S. Park, W. W. Chu, J. Yoon, and C. Hsu. Efficient searches
for similar subsequences of different lengths in sequence databases. In
Proceedings of the IEEE ICDE, 2000.
[PRC94] P. Bieganski, J. Riedl, and J. V. Carlis. Generalized suffix
trees for biological sequence data: Applications and implementation. In
Proceedings of the International Conference on System Sciences, 1994.
[RJ93] L. Rabiner and B. H. Juang. Fundamentals of Speech Recognition.
Prentice Hall, 1993.
[Ste94] G. A. Stephen. String Searching Algorithms. World Scientific
Publishing, 1994.
[SYV97] A. Prasad Sistla, Clement Yu, and Raghu Venkatasubrahmanian.
Similarity based retrieval of videos. In Proceedings of the 13th
International Conference on Data Engineering, Birmingham, UK, April
1997. IEEE, IEEE Computer Society Press.
[YJF98] B. K. Yi, H. V. Jagadish, and C. Faloutsos. Efficient retrieval
of similar time sequences under time warping. In Proceedings of the
IEEE ICDE, 1998.
[YO96] N. Yazdani and M. Ozsoyouglu. Sequence matching of images. In
Proceedings of the SSDBM, 1996.