ASO PCR
advertisement
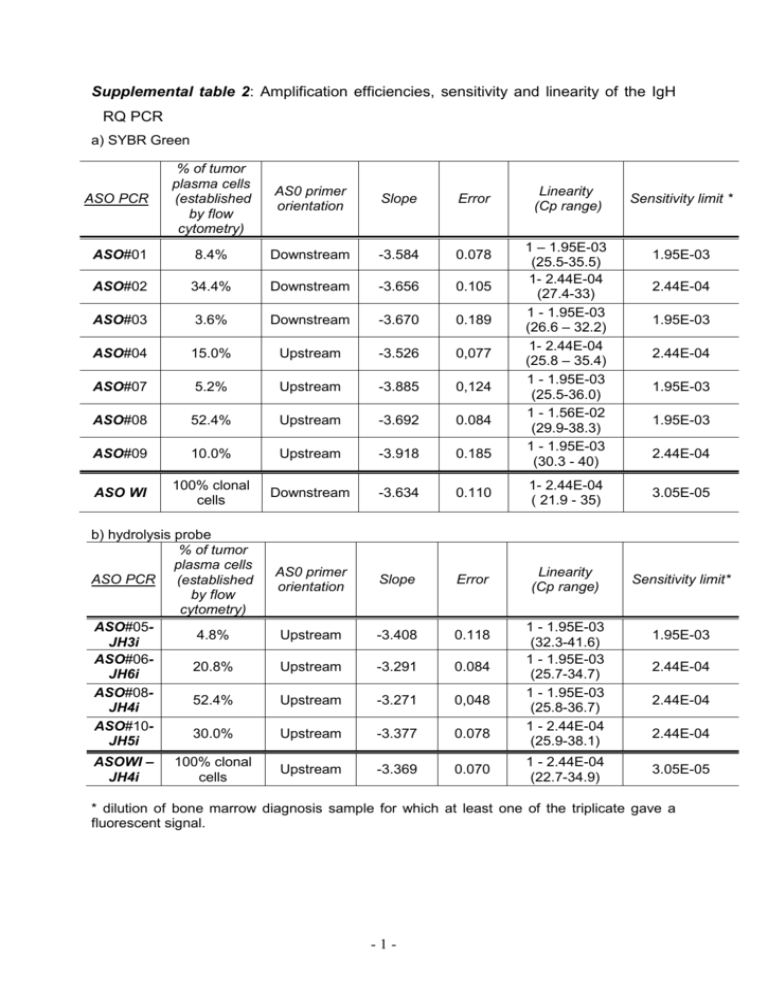
Supplemental table 2: Amplification efficiencies, sensitivity and linearity of the IgH RQ PCR a) SYBR Green ASO PCR % of tumor plasma cells (established by flow cytometry) AS0 primer orientation Slope Error ASO#01 8.4% Downstream -3.584 0.078 ASO#02 34.4% Downstream -3.656 0.105 ASO#03 3.6% Downstream -3.670 0.189 ASO#04 15.0% Upstream -3.526 0,077 ASO#07 5.2% Upstream -3.885 0,124 ASO#08 52.4% Upstream -3.692 0.084 ASO#09 10.0% Upstream -3.918 0.185 ASO WI 100% clonal cells Downstream -3.634 0.110 1- 2.44E-04 ( 21.9 - 35) 3.05E-05 AS0 primer orientation Slope Error Linearity (Cp range) Sensitivity limit* Upstream -3.408 0.118 Upstream -3.291 0.084 Upstream -3.271 0,048 Upstream -3.377 0.078 Upstream -3.369 0.070 b) hydrolysis probe % of tumor plasma cells ASO PCR (established by flow cytometry) ASO#054.8% JH3i ASO#0620.8% JH6i ASO#0852.4% JH4i ASO#1030.0% JH5i ASOWI – JH4i 100% clonal cells Linearity (Cp range) 1 – 1.95E-03 (25.5-35.5) 1- 2.44E-04 (27.4-33) 1 - 1.95E-03 (26.6 – 32.2) 1- 2.44E-04 (25.8 – 35.4) 1 - 1.95E-03 (25.5-36.0) 1 - 1.56E-02 (29.9-38.3) 1 - 1.95E-03 (30.3 - 40) 1 - 1.95E-03 (32.3-41.6) 1 - 1.95E-03 (25.7-34.7) 1 - 1.95E-03 (25.8-36.7) 1 - 2.44E-04 (25.9-38.1) 1 - 2.44E-04 (22.7-34.9) Sensitivity limit * 1.95E-03 2.44E-04 1.95E-03 2.44E-04 1.95E-03 1.95E-03 2.44E-04 1.95E-03 2.44E-04 2.44E-04 2.44E-04 3.05E-05 * dilution of bone marrow diagnosis sample for which at least one of the triplicate gave a fluorescent signal. -1- Efficiency determination Slopes from the standard curve, permitted to determine the efficiencies of ASO PCR. The best efficiency is achieved when doubling the PCR specific product at each cycle resulting in an efficiency of 2 and a slope of -3.32 according to the equation E=10-1/slope. Slopes between -4.0 and -3.2 which corresponds to an efficiency varying between 89% and 100%, are considered as acceptable. Upstream versus downstream ASO IgH SYBR Green quantitative PCR ASO IgH PCRs were adapted for SYBR Green I detection system. Successful amplifications were obtained using the Qiagen SYBR Green kit either with the downstream oriented ASO primer for 4 ASO PCRs or with the upstream oriented ASO primer for 4 other ASO PCRs (Supplemental table 2a). For SYBR Green I optimized ASO PCR reactions, an acceptable PCR efficiency was obtained with slopes ranging from -3.526 to -3.918. It has been reported that smaller amplicons were more efficiently amplified. Howewer, we did not reach the same conclusion as the efficiencies between ASO downstream (PCR product from about 250 bp) and upstream strategies (PCR product about 80 bp) were comparable. Therefore, both ASO primer orientations can be recommended. ASO RQ PCR with SYBR Green versus consensus hydrolysis probes approaches For SYBR Green I detection, the slopes examined for 8 ASO PCRs ranged from 3.526 to -3.918, corresponding to a mean value of -3.704 with an error ≤ to 0.189 (Supplemental table 2a). For hydrolysis probe detection, the slope mean of 5 ASO PCRs, was higher and attained -3.337 (ranging from -3.271 to -3.408) with an error ≤ to 0.118 (Supplemental table 2b). These results indicated that hydrolysis probe detection format led to a better PCR efficiency. For the MM ASO PCR, the sensitivity limit varied between 1.95E-03 and 2.44E-04 and was reduced compared to the WI cell line. The reduced sensitivity limit for ASO #01, #03 and #07 was explained by a weak initial tumor load inferior to 10% of total MNC as established by flow cytometry. For hydrolysis probe format, the sensitivity limit reached 2.44E-04 except for the ASO #05 PCR which only attained 1.95E-03 because of the low tumor burden (4,8%) in the BM sample DNA from diagnosis. For ASO #08 PCR analysed by both methods, the linearity and the sensitivity limit were reduced with the SYBR Green I detection approach, therefore the hydrolysis probe detection was advised and used for the residual tumor cells follow-up when possible. -2-










