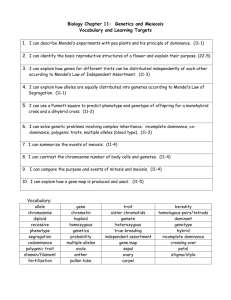
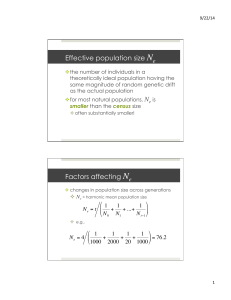
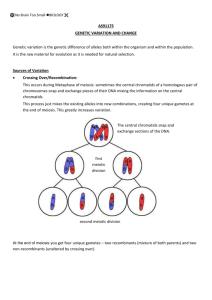
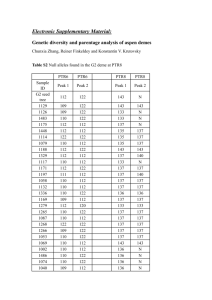
loci evolution
advertisement

B1.4.
Research method (TWO A4 PAGES)
For each objective explain the methodological approach that will be employed in the project and justify it in
relation to the overall project objectives. When any novel methods or techniques are proposed, explain why the
approach is likely to succeed.
1. What is genetic drift consequence on several loci?
In reality, populations are always finite and may be small enough for chance
fluctuations due to random drift to be quite important. In order to study the
consequences of drift on evolution, a wide body of theories was developed,
initially by Fisher (1930) and Wright (1931).
The simplest Wright-Fisher model describes the evolution of a two alleles locus
(A1/A2) in a diploid population of constant size N, undergoing random mating
(and with non-overlapping generations). This is the "random drift model" of
population genetics. The evolution of the random variable X(t+1) describing the
number of copy of one of the alleles, say A1, present in the population at
generation t+1 is fully described by a Markov chain. Given X(t) the number of A1
alleles in the generation t, X(t+1) follows a binomial law B(X(t)/2N,2N). The
process {X(t), t = 0, 1, ..., n} is an example of a time homogeneous Markov
chain. It has a binomial transition matrix T = pij, and a state space S = {0, 1,
..., 2N} with two absorbing states 0 and 2N. The properties of this Markov chain
have been extensively studied. The first four moments of the distribution, the
probability of fixation and the rate of decrease in heterozygosity have been
derived (refs), as well as the duration time to fixation that were computed
using diffusion approximation (refs). The theory was further extended to take
into account several alleles at one locus (refs). The two loci-two alleles case
was also studied in details and extension to more alleles were considered. In
these cases, the hidden assumptions that gametes are drawn at random from an
infinite pool, and as a consequence that all the genotypic combinations can be
formed, are justified.
The number of gametes that can be formed at K loci each with (a) alleles
increases exponentially with K. More precisely, it is a^K. The number of
possible genotypes is the number of pairs of gametes a^K(a^K+1)/2. We want to
study the cases where the size of the population is smaller than the total
number of genotypes that are possible. In this case, not all the possible
genotypes will be present in the population. As a result, some of the gamete
combinations might not be possible to achieve. In this case, the assumption that
genetic drift can be modelled by random sampling of gametes from an infinite
pool cannot hold. The Wright-Fisher model must thus be modified to address the
multiloci-multialleles case.
To take in account this drawback, we propose to consider a haploid population of
constant size N undergoing random mating. This time, given population state at
generation t, the next generation is formed as following: we first sample
randomly N pairs of parents and then constitute N individuals by randomly
sampling one of the two parent’s alleles at each locus.
We will try to obtain a complete statistical multiloci description of the
evolution of this model population permitting for example to compute the
multiloci fixation probability or the distribution of the inbreeding
coefficient.
2. Neutrality tests
The methods used to detect the footprints of selection at the molecular level
are based on two principles. Firstly, the data concern a candidate gene that is
either directly affected by selection (unfrequent case) or a locus that is in
linkage disequilibrium with the gene or the genomic region under selection (more
frequent case). Secondly, selection on the candidate or on the selected genomic
region produces a departure from the prediction based on a neutral assumption.
As a result, any data exhibiting significant departure from the neutral
hypothesis indicate the footprint of selection. This can be done for example by
studying the local reduction of molecular diversity around a selected region
(selective sweep, Maynard-Smith and Haigh 1974 ; Kaplan et al. 1989; Begun and
Aquadro 1992), or by measuring a test statistics (for example FST, Wright 1969)
like the FST for a set of markers and by looking for outliers i.e. the markers
for which the test statistics deviates significantly from the distribution
expected under the hypothesis of genetic drift (Beaumont and Nichols 1996 ;
Flint et al. 1999 ; Vigouroux et al. 2002 ; Vitalis et al. 2003).
A terminer.
3. Selection