Electronic Supplementary Material
advertisement

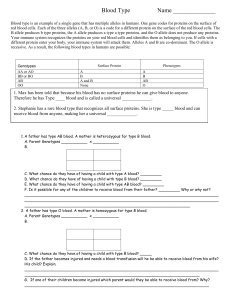
Electronic Supplementary Material: Genetic diversity and parentage analysis of aspen demes Chunxia Zhang, Reiner Finkeldey and Konstantin V. Krutovsky Table S2 Null alleles found in the G2 deme at PTR8 Sample ID G2 seed tree 1129 1126 1483 1175 1448 1114 1079 1188 1329 1117 1171 1197 1058 1132 1336 1169 1279 1265 1087 1268 1266 1053 1069 1002 1486 1074 1040 PTR6 PTR6 PTR8 PTR8 Peak 1 Peak 2 Peak 1 Peak 2 112 122 143 N 109 109 110 112 112 122 110 112 112 110 112 111 110 110 110 109 112 110 110 122 109 110 110 110 110 110 109 122 122 122 112 112 122 112 122 112 112 122 112 112 112 122 112 120 122 112 122 122 122 112 112 122 122 112 143 133 133 137 135 135 135 143 137 133 137 137 137 137 136 137 133 137 137 137 137 137 143 136 136 136 136 143 N N N 137 137 137 143 140 N 137 140 137 137 136 137 133 137 137 137 137 137 143 N N N N 1205 110 122 137 137 1490 122 122 139 N 1222 112 113 133 N Note: N refers to null alleles. A microsatellite null allele is defined as any allele at a microsatellite locus that consistently fails to amplify to detected levels via the polymerase chain reaction (PCR). In our study, we detected the null alleles by comparing the genotypes of maternal genotypes and their progenies’s genotypes, we found some null alleles at the loci PTR8 in the G2 deme. The seed tree of the G2 deme should have maternal allele 137 inferred from the progenies’ genotypes, but we did not detect it in our PCR amplification. Some progenies of the G2 seed tree with the homozygote genotype at PTR8, such as 136136, 139139, 133133 had no maternal alleles 143.