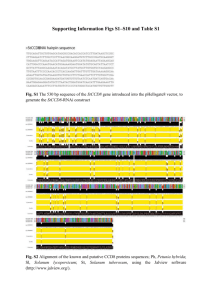
Supplementary Table 1: Laboratory protocols used in this study
advertisement

Supplementary Table 1: Laboratory protocols used in this study PCR conditions 2 PCR product analysis Sequencing Invitrogen 2μl of the cDNA Super-script mixture II 1 Taq (Roche) 94oC, 3 min Agarose gel 35 cycles of 94oC, 30s 60oC, 30s 72oC, 45c then 72oC, 5 min Sequencing of cDNA pool using ABI3730xl Random hexamers Invitrogen 2μl of the cDNA Super-script mixture II 1 GoTaq (Promega) 94oC, 2 min 3 cycles of 94oC, 30 s xoC, 40 s 72oC, 1 min 3 cycles of 94oC, 30 s (x-2)oC, 40 s 72oC, 1 min 30 cycles of 94oC, 30 s (x-4)oC, 40 s 72oC, 1 min then 72oC, 7 min Agilent or Qiaxell automated electrophoresis or agarose gel and gel purification of aberrant bands (QIAquick) Sequencing of cDNA pool or purified bands using ABI3730xl 1µg Random hexamers Invitrogen Superscript III; 50°C for 60 min 95°C 8 min 38 cycles of 94°C 40 s x°C 45 s 72°C 45 s; then 72°C 10 min Agarose gel and gel purification of aberrant bands (QIAquick) and/or DHPLC "doublestrand multiple fragments" application Sequencing of cDNA purified bands using ABI3130 N/A 0.5-1µg Random hexamers Applied 1l of the cDNA Biosystems mixture Multiscribe 1 Phusion (Finnzymes) Agarose gel, PCR98C 30 s Blunt-TOPO cloning 35 cycles of 98C 15 s 56C 20 s 72C 30 s then 72C 5 min Sequencing of cDNA purified bands using ABI3730 RNeasy Kit and RNaseFree DNase I1(Qiagen) 1µg Random Applied hexamers Biosystems and oligo dT High Capacity RNA-tocDNA Master Mix 1 94°C 5 min, 35 cycles of 94°C 30 s x°C 30 s 72°C 30 s 72°C 7 min Sequencing of cDNA pool using ABI3130xl Laboratory acronym Cells / tissue (collection method) Culture conditions Nonsense mediated decay inhibition RNA extraction method Dnase 1 treatment Amount of cDNA total RNA synthesis used in cDNA primer synthesis cDNA synthesis protocol OUH LCL LCL + RPMI + 10% serum + 1% Pen/strep With and without puromycin 200μg /ml for 2 hours Qiagen RNeasy Minikit 1 RNase-Free DNase I1(Qiagen) 3µg Gene specific FPGMX Fresh blood (EDTA tubes or Heparin tubes for PHA) 1ml blood + 9 ml RPMI + 20% serum + 1% Pen/strep + PHA (100μg/ ml) 72 hrs With and without puromycin 200μg /ml for 8 hours Qiagen RNeasy Minikit 1 RNase-Free DNase I1(Qiagen) 1µg IOV LCL LCL + RPMI + 13% serum + 2mM lglutamine None Qiagen RNeasy Minikit 1 RNase-Free DNase I1(Qiagen) CBCS Fresh blood N/A (EDTA or Citrate tubes) N/A TriZOL HVH Fresh blood N/A (EDTA tubes) N/A TriZOL and RNA cleanup with Qiagen RNeasy Mini Kit PCR amplification template 3μl of the cDNA mixture AmpliTaq gold (Applied Biosystems) 2μl of the cDNA mixture EcoTaq (Ecogen) Agarose gel 95oC, 2 min Agarose gel 38 cycles of 95oC, 30 s 56oC, 30 s 72oC, 60 s then 72oC 2 min AAS Fresh blood N/A (EDTA tubes) N/A TriZOL N/A 1-5µg Random hexamers Invitrogen Superscript III1 2μl of the cDNA mixture AmpliTaq (Applied Biosystems) ICO Fresh blood 500 μL PBL (EDTA + 7mL RPMI tubes) + 10% serum + 1% Pen/strep + PHA (10 g per ml RPMI medium) 72-120 hrs Puromycin 250g/ml for 4-6 hours TriZOL N/A 0.5-1µg Random hexamers Invitrogen Superscript II 1 2μl of the cDNA 96oC 5 min mixture 35 cycles of MegaMix (Microzone) 96oC 30s-60 s xoC 30s-60 s 72oC 60-120s then 72oC, 10 min 1 According to manufacturers' protocols 2 x=melting temperature depending on the pair of primers used in the reaction LCL: lymphoblastoid cell lines Pen/strep: penicillin-streptomycin PHA: phytohaemaglutinin Agarose gel Sequencing of cDNA pool using ABI310xl, allele-specific sequencing Sequencing of cDNA pool using ABI3130 Supplementary Table 2: Methodology and primer sequences used to assay variants GENE VARIANT (HGVS) Exon /Intron Location of Variant Method Group BRCA1 c.301+6T>C RT-PCR, sequencing, DHPLC IOV BRCA1 BRCA1 c.441+2T>A c.548-8delT I6 I7 I8 BRCA1 c.4097-15T>C I11 BRCA1 c.4184_4185 +2del E12 I12 BRCA1 c.4357+1G>A I13 BRCA1 c.4358-4delA I13 BRCA1 c.4987-2A>G I16 RT-PCR, sequencing RT-PCR, sequencing RT-PCR, sequencing RT-PCR, sequencing RT-PCR, sequencing RT-PCR, sequencing RT-PCR, sequencing cDNA primers Forward PCR primers (5’-3’) Location of Forward Primer Reverse PCR primers (5’-3’) Location of Reverse Primer Expected band size (bp) random hexamers CAAGGAACCTGTCTCCACAAAGTG exon 3 AAGTCTTTTGGCACGGTTTCTG exon 7 319 random hexamers CAAGGAACCTGTCTCCACAAAGTG exon 3 CGTCTTTTGAGGTTGTATCCGCTG exon 8 436 random hexamers GAAAGGGCCTTCACAGTGTC exon 5 TCTTTTGGCACGGTTTCTGT exon 7 244 CAGAAGAAAGGGCCTTCACA exon 5 TTTGGCATTATCAACTGGCTTATC exon 11 2670 CAGCTTGACACAGGTTTGGA exon 6** GATCAGCATTCAGATCTACC exon 11** 757 TGCAAGTTTG AAACAGAAC TAACCTTGGAACTGTGAGAAC exon 8 CAGAATCCAAACTGATTTCATC exon 10 192 TGCAAGTTTG AAACAGAAC AAAACTTTTATTGATTTATTTTTTGG intron 8 CAGAATCCAAACTGATTTCATC exon 10 155 if intron retention TGCAAGTTTG AAACAGAAC AAATATTTCTAGTTGAATATCTG intron 8 CAGAATCCAAACTGATTTCATC exon 10 217 if intron retention random hexamers ACCGTTGCTACCGAGTGTCT exon 11 CCCTGCTCACACTTTCTTCC exon 16 1168 CAAAAGCGTCCAGAAAGGAG exon 11 CCCTGCTCACACTTTCTTCC exon 16 1370 ACCGTTGCTACCGAGTGTCT exon 11** CTGTTGCTCCTCCACATCAA exon 15** 879 random hexamers GAAAATAATCAAGAAGAGCAAAGCA exon 11 CCCTGCTCACACTTTCTTCC exon 16 847 GGCAAGTAAG ATGTTTC CTGCGAAATCCAGAACAAAG exon 13 TGTTGCTCCTCCACATCAAC exon 15 290 GGCAAGTAAG ATGTTTC GTAGATTTGTTTTCTCATTCCA intron 13 TGTTGCTCCTCCACATCAAC exon 15 285 if intron retention random hexamers GGCCTTTCTGCTGACAAGTT exon 14 ATTCTCTTGCTCGCTTTGGA exon 20 853 FPGMX random hexamers OUH FPGMX FPGMX random hexamers FPGMX OUH FPGMX Sequencing primers, if different from PCR primers (5’-3’) CAGCTTGACACAGGTTTGGA (forward exon 6) GATCAGCATTCAGATCTACC (reverse, exon 11) CTGTTGCTCCTCCACATCAA (reverse, exon 15) ACCGTTGCTACCGAGTGTCT (forward, exon 11) CTGTTGCTCCTCCACATCAA (reverse, exon 15) AAGACAGAGCCCCAGAGTCA (forward exon 16) AAGACAGAGCCCCAGAGTCA exon 16** GACCTTGGTGGTTTCTTCCA exon 20** 504 BRCA1 c.5074G>C E17 RT-PCR, sequencing IOV random hexamers CCCCAGAAGAATTTATGCTCGTG exon 16/17 TTCTCTTGCTCGCTTTGGACC exon 20 290 BRCA1 c.5075-107A>G I17 RT-PCR, sequencing ICO random hexamers GGGTCAACAAAAGAATGTCCA exon 16 TCTTCCATTGACCACATCTCC exon 20 300 BRCA1 c.5333A>G E22 ICO random hexamers GTTTGCCAGAAAACACCACA exon 17 CACAGGTGCCTCACACATCT exon 24 496 FPGMX random hexamers TGAAGTCAGAGGAGATGTGG exon 20 TGGTAGAGTGCTACACTGTC exon 24 338 RT-PCR, sequencing BRCA2 c.3G>A E2 RT-PCR, sequencing HVH random hexamers and anchoredoligo (dT)18 AGCGTGAGGGGACAGATTT exon 1 AGTCAGCCCTTGCTCTTTGA exon 3 380 BRCA2 c.316+5G>A I3 RT-PCR, sequencing AAS random hexamers AAGAGAGGCCAACATTTTTTGAAA exon 2 GGTGTCTGACGACCCTTCACA exon 6/7 514 BRCA2 c.425+33A>G I4 ICO random hexamers TCAGCTGGCTTCAACTCCAATAA exon 3 GAACTAAGGGTGGGTGGTGTAGC exon 7 402 HVH random hexamers and anchoredoligo (dT)18 GGATCCAAAGAGAGGCCAAC exon 2 TGAAACAAACTCCCACATACCA exon 6 488 GATCACGGATCCAAATATGTTTTGGTG TCTGACGACC exon 7 GATCACGGATCCAAATATGTTTTGGTG TCTGACGACC exon 7 RT-PCR, sequencing c.426-22G>T I4 minigene* CBCS N/A GATCACGAATTCTCCAGCAGCTGAA ATTTGTGAGTAC intron 4 BRCA2 c.516+18T>C I6 minigene* CBCS N/A GATCACGAATTCTCCAGCAGCTGAA ATTTGTGAGTAC intron 4 BRCA2 BRCA2 c.632-16A>C I7 RT-PCR, sequencing HVH random hexamers and anchoredoligo (dT)18 CCGCTGTACCAATCTCCTGT exon 3 TTCCAATGTGGTCTTTGCAG exon 10 589 BRCA2 c.992A>T E10 RT-PCR, sequencing ICO random hexamers TCAAAGAGAAGCTGCAAGTCA exon 9 TTCCTCAGAATTGTCCCAAAA exon 11 1236 BRCA2 c.1096T>G E10 RT-PCR, sequencing FPGMX random hexamers CAAATCAAAGAGAAGCTGCAA exon 9 ACCTTTGAGCTTGTCTGACA exon 11 1634 FPGMX random hexamers CAAATCAAAGAGAAGCTGCAA exon 9 ACCTTTGAGCTTGTCTGACA exon 11 1634 FPGMX random hexamers CAGTGGCTTCTTCATTTCAGG exon 10 GGAGTGCTTTTTGAAGCCTTT exon 12 9282 BRCA2 c.1788T>C E10 RT-PCR, sequencing BRCA2 c.3156A>C E11 RT-PCR, sequencing GACCTTGGTGGTTTCTTCCA (reverse, exon 20) TCTGGAGCGGACTTATTTACCAAA TCTGGAGCGGACTTATTTACCAAG 648 pSPL3 vector-specific primers: 5’-TCTGAGTCACCTGGACAACC-3’ 5’-ATCTCAGTGGTATTTGTGAGC-3’ 648 pSPL3 vector-specific primers: 5’-TCTGAGTCACCTGGACAACC-3’ 5’-ATCTCAGTGGTATTTGTGAGC-3’ CAGCCACCACCACACAGA (forward, exon 10) TCTGGTTTTCAGGCACTTCA (reverse, exon 11) CCAATTTCAAATCACAGTTTTGGAGGT (forward, exon 11) GGAGTGCTTTTTGAAGCCTTT (reverse, exon 12) BRCA2 c.7397C>T E14 RT-PCR, sequencing BRCA2 c.7435+6G>A I14 RT-PCR, sequencing HVH BRCA2 c.8421G>T E19 RT-PCR, sequencing FPGMX BRCA2 c.8754+3G>C E21 RT_PCR, sequencing minigene* HVH random hexamers and anchoredoligo (dT)18 random hexamers and anchoredoligo (dT)18 AGGCTTCAAAAAGCACTCCA exon 12 TCCACCATCAGCCAACTGTA exon 16 842 AGGCTTCAAAAAGCACTCCA exon 12 TCCACCATCAGCCAACTGTA exon 16 842 random hexamers ATGGAAAGGGATGACACAGC exon 18 CTGATGATGGACGCCAAATA exon 23 956 CBCS random hexamers CGGCCTGCTCGCTGGTAT exon 19 GCCTTCCTAATTTCCAACTGGATCTG exon 22 503 CBCS N/A GATCACGAATTCTTCCTGGAAAACTT ATAGCA intron 20 GATCACCTCGAGTTAGGGTAGAGGAT TATCAAGTACA intron 21 CTAACAGTACTCGGCCTGCTC (forward, exon 19) CAGATTCCATGGCCTTCCTA (reverse, exon 22) pSPL3 vector-specific primers: 5’-TCTGAGTCACCTGGACAACC-3’ 5’-ATCTCAGTGGTATTTGTGAGC-3’ * Minigene primers listed in the body of the table are those used for the cloning of the specific exons into the minigene vector. The following vector-specific primers were used for the final PCR: Forward primer: 5GTGAACTGCACTGTGACAAGCTGC-3; Reverse primer: 5-CACCTGAGGAGTGAATTGGTCG-3 **Primers used for a nested PCR Supplementary Table 3: Bioinformatic prediction of cryptic splice site usage for variants associated with splicing aberrations in vitro* Score for variant sequence Gene Variant Site SSF MaxEnt NNSPLICE GeneSplicer [0-100] [0-12] [0-1] [0-15] ≥70 ≥0 ≥0.4 ≥0 71.44 4.65 — — 74.93 5.63 0.65 — 73.84 1.23 — — 72.36 3.77 0.54 — 79.7 — — — — 4.78 0.63 2.9 78.16 5.73 0.57 — Exon 12 - c.4112 — 0.35 — — Exon 12 - c.4114 — 0.22 — — Intron 12 - c.4185+9 — 1.63 — — — 1.3 — — Intron 12 - c.4185+29 — 0.29 — — Intron 12 - c.4185+31 — 1.3 — — Intron 12 - c.4185+57 — 3.71 — 0.49# Intron 12 - c.4185+78 70.04 1.69 — — 73.58 5.42 0.89 — Intron 16 - c.4987-88 70.68 2.73 — — Intron 16 - c.4987-39 — 0.42 — — 72.04 — — — Exon 17 - c.5048 73.68 — — — Intron 17 - c.5074+1 75.96 4.36 — 3.95 78.13 — — — Predicted location of alternative splice site Location of cryptic splice site(s) utilized in vitro Exon 6 - c.226 BRCA1 c.301+6T>C D Exon 6 - c.292 Exon 6 - c.292 Intron 6 - c.301+106 Exon 7 - c.379 D BRCA1 c.441+2T>A Intron 7 - c.441+91 A Exon 7 - c.379 (donor); Exon 8 - c.445 (acceptor) Exon 8 - c.445** Intron 11 - c.4097-1 BRCA1 BRCA1 BRCA1 BRCA1 c.4184_4185+2del c.4357+1G>A c.4987-2A>G c.5074G>C D D A D Intron 12 - c.4185+11 Intron 13 - c.4357+71 Intron 16 - c.4987-37 Intron 17 - c.5074+55 None - exon 12 skipping None - exon 13 skipping None - exon 17 skipping Intron 17 - c.5074+153 Intron 17 - c.5074+60 BRCA2 c.316+5G>A D 79.17 — — — Intron 17 - c.5074+153** — 0.56 — 1.62 Intron 3 - c.316+17 — 1.91 — — 74.47 — — — 71.17 — — — — 2.69 — — 73.27 5.73 0.57 # — Intron 21 - c.8754+46 90 8.68 1 6.64 Intron 21 - c.8754+87 — 4.82 — — Intron 3 - c.316+51 None - exon 3 skipping Intron 3 - c.316+100 Exon 21 - c.8682 Intron 21 - c.8754+8 BRCA2 c.8754+3G>C D Intron 21 - c.8754+46 * Predictions for cryptic sites consistent with the observed in vitro results are noted in bold. – No predicted site. D = Donor site A = Acceptor site ** Acceptor cryptic splice site location falls outside the region analyzed, input sequence increased on the basis on results from RT-PCR analysis to generate scores shown in Table. # Score for alternative splice site was not generated for wildtype sequence input. Supplementary Table 4: Posterior Probability of Variant Pathogenicity as estimated using Multifactorial Likelihood Analysis Likelihood ratios derived from analysis of a Myriad dataset** Gene Variant (HGVS nomenclat ure) Intron-Exon Location (missense AGVGD score) BRCA1 c.301+6T>C Intron 6 c.441+2T> A Intron 7 within splice consensus site BRCA1 BRCA1 BRCA1 BRCA1 BRCA1 BRCA1 c.5488delT c.409715T>C c.4184_418 5+2del c.4357+1G >A c.43584delA Intron 8 Intron 11 Exon12Intron 12 spanning splice consensus site Total # LLR LLR based Observatio based on coLLR based on Total LLR Total LR from ns of the Prior on occurrence Cosegregatio from Myriad Myriad variant in Probability* report of with a n data information information the Family deleterious combined History mutation datasets Instances of cooccurrence reported by ENIGMA sites Frequency of mutations in relevant gene, in reporting laboratory (p1) LR based on cooccurrence with a deleterious mutation ENIGMA cosegregatio n pedigrees available LR based on cosegregation data Odds for Causality (all LRs combined) Posterior Odds of a variant being deleterious Posterior Probability of a variant being deleterious Multifactorial Likelihood Classification according to the IARC 5 Class system^ 0.0021221 6 0.0007456 2 0.0007450 7 Class 1 Not Pathogenic / Low Clinical Significance 1.0475142 9 1.0338607 0 3.6034491 4 25.140342 86 0.3632483 5 1.2660767 3 0.9617449 5 0.2664579 4 0.5587086 8 Class 4 Likely Pathogenic Class 3 Uncertain Class 3 Uncertain 0.26 ND ND ND ND 1 1 0 0.058 1.061 1 0.002 0.96 ND ND ND ND 1 1 0 0.045 1.048 0 1 0.26 ND ND ND ND 1 1 0 0.033 1.034 0 1 0.26 ND ND ND ND 1 1 0 0.045 1.048 1 3.44 0.96 ND ND ND ND 1 1 0 0.045 1.048 1 2.09 0.96 6.09 0.6 ND 6.68 4786300.9 23 1 0 0.045 1.048 0 1 0.26 ND ND ND ND 1 1 0 0.033 1.034 0 1 Intron 13 within splice consensus site Intron 13 Likelihood ratios derived from analysis of ENIGMA datasets*** 2.1893048 6 52.543316 57 0.9813235 3 Class 4 Likely Pathogenic 5013718.5 9 1.0338607 0 120329246 .23 0.3632483 5 0.9999999 9 0.2664579 4 Class 5 Pathogenic Class 3 Uncertain Classification based on Combined Interpretation of Multifactorial Analysis, Frequency Data and Splicing Results Class 1 Not Pathogenic / Low Clinical Significance Class 5 Pathogenic (observed splicing aberration) Class 3 Uncertain Class 3 Uncertain Class 5 Pathogenic (observed splicing aberration) Class 5 Pathogenic (multifactorial, observed splicing aberration) Class 3 Uncertain BRCA1 BRCA1 c.49872A>G c.5074G>C p.Asp1692 His Intron 16 within splice consensus site c.5075107A>G BRCA1 c.5333A>G p.Asp1778 Gly Exon 22 (C0) c.3G>A p.Met1Ile Exon 2, disrupts start codon BRCA2 Intron 17 BRCA2 c.316+5G> A Intron 3 BRCA2 c.425+33A >G Intron 4 BRCA2 BRCA2 BRCA2 BRCA2 c.42622G>T c.516+18T> C c.63216A>C c.992A>T p.Lys331Ile Intron 4 Intron 6 Intron 7 BRCA2 BRCA2 BRCA2 BRCA2 c.1096T>G p.Leu366V al c.1788T>C p.=? c.3156A>C p.=? 0.9617449 5 Class 4 Likely Pathogenic 1 1.1291401 8 1.0627528 8 4.8137028 8 0.3733996 6 0.8279925 9 0.2718798 3 Class 3 Uncertain Class 3 Uncertain 0 1 0.3337152 9 0.0033708 6 0.0033595 4 Class 2 Likely Not Pathogenic Class 2 Likely Not Pathogenic 1.072 1 2.00 1.028 1 3.16 2.1445923 9 5.6436016 7 51.470217 39 1.9828870 7 0.9809415 7 0.6647543 2 Class 4 Likely Pathogenic Class 3 Uncertain Class 4 Likely Pathogenic Class 4 Likely Pathogenic 0.0054650 9 0.3597162 0 0.3597162 0 0.0946360 0 0.0054353 8 0.2645524 1 0.2645524 1 0.0864543 1 Class 2 Likely Not Pathogenic Class 3 Uncertain Class 3 Uncertain Class 3 Uncertain Class 2 Likely Not Pathogenic Class 3 Uncertain Class 3 Uncertain Class 3 Uncertain ND ND ND ND 1 1 0 0.045 1.048 0 1 0.81 -0.045 0.073 ND 0.027 1.0641430 18 1 0 0.058 1.061 0 1 0.26 ND ND ND ND 1 2 0× 0.030 1.063 0 0.3090295 43 2 0 0.030; 0.045 1.080 1 0 0.068 1 0.028 0.041; 0.026 1.140 2 0.01 0.01 -0.64 0.13 ND -0.51 0.96 ND ND ND ND 0.26 0.18 0.06 ND 0.24 1 1.7378008 29 0.26 ND ND ND ND 1 4 0 1, in cis (BRCA2 c.9018C>A p.Tyr3006X ) 0.26 ND ND ND ND 1 1 0 0.024 1.024 0 1 1 0 0.024 1.024 0 1 1 0 0.068 1.072 0 1 0.0155544 8 1.0238076 5 1.0238076 5 0.2693486 3 1 0.8561451 8 0.0086479 3 0.0085737 9 Class 2 Likely Not Pathogenic Class 2 Likely Not Pathogenic 1 1.0252128 3 0.0103556 9 0.0102495 4 Class 2 Likely Not Pathogenic Class 2 Likely Not Pathogenic 1 1.0252128 3 0.0103556 9 0.0102495 4 Class 2 Likely Not Pathogenic Class 1 Not Pathogenic / Low Clinical Significance 0.26 ND ND ND ND 0.26 -0.65 0.05 ND -0.6 1 0.2511886 43 -0.085 0.8222426 5 Exon 10 (C0) 0.01 BRCA2 25.140342 86 0.96 Exon 17 (C65) BRCA1 1.0475142 9 Class 5 Pathogenic (observed splicing aberration) Class 5 Pathogenic (observed splicing aberration) Class 3 Uncertain -0.096 0.01 ND 1 0 0.041 1.041 0 Exon 10 (C0) 0.01 Exon 10, silent with low probability to disrupt splicing Exon 11, silent with low probability to disrupt splicing c.7397C>T p.Ala2466V al Exon 14 (C0) c.7435+6G >A Intron 14 0.01 0.01 0.01 0.26 ND ND ND ND -0.97 ND ND ND ND 0.46 ND ND ND ND -1.06 ND ND ND 1 1 1 ND 1 -1.56 0.0275422 87 1 1 1 4 1 0 0 0.026 0.026 0 0.026 0 0.068; 0.026 0 0.068 1.025 1.025 1.025 1.150 1.072 0 0 1 2 0 0.025 0.0256303 2 0.0002588 9 0.0002588 3 Class 1 Not Pathogenic / Low Clinical Significance 0.97 1.1114151 7 0.0112264 2 0.0111017 8 Class 2 Likely Not Pathogenic Class 1 Not Pathogenic / Low Clinical Significance Class 1 Not Pathogenic / Low Clinical Significance 1 0.0295334 9 0.0103766 3 0.0102700 6 Class 2 Likely Not Pathogenic Class 2 Likely Not Pathogenic BRCA2 BRCA2 c.8421G>T p.=? c.8754+3G >C Exon 19, silent with inconsistent prediction to create a de novo acceptor site Intron 21 0.01 ND ND ND ND 1 1 0 0.026 1.025 0 1 0.26 ND ND ND ND 1 1 0 0.024 1.024 0 1 1.0252128 3 1.0238076 5 0.0103556 9 0.3597162 0 0.0102495 4 0.2645524 1 Class 2 Likely Not Pathogenic Class 3 Uncertain Class 2 Likely Not Pathogenic Class 4 Likely Pathogenic *Prior Probability based on location of variant relative to splice junction (intronic variants, Easton et al 2007) or physicochemical characteristics of encoded missense substitution (exonic variants, Tavtigian et al 2008). ** Log likelihood ratio (LLR) estimates derived from analysis of 70,000 BRCA1 and BRCA2 tests as reported in Easton et al 2007. ***Mutation detection rate for relevant laboratory reporting the variant; co-occurrence likelihood ratios were calculated separately where two laboratories detected the same variant. ^Classifications as described in Plon et al (Plon, et al., 2008) × co-occurrence with mutation in BRCA2