Fundamentals of Molecular Evolution: Problems Practical
advertisement
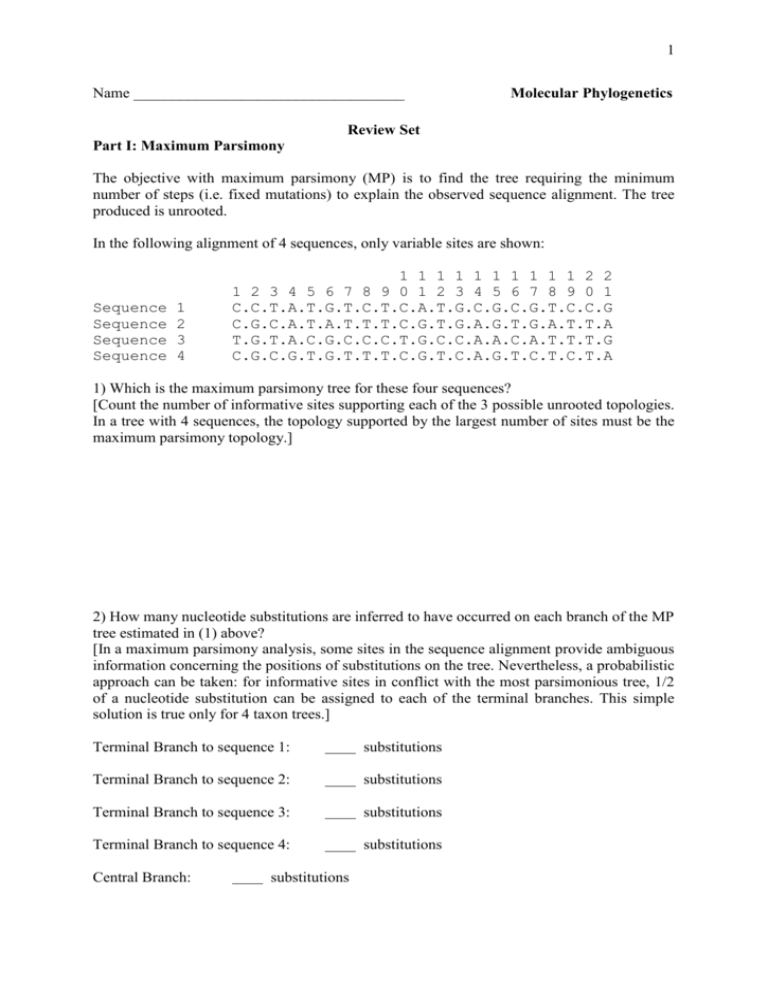
1 Name ___________________________________ Molecular Phylogenetics Review Set Part I: Maximum Parsimony The objective with maximum parsimony (MP) is to find the tree requiring the minimum number of steps (i.e. fixed mutations) to explain the observed sequence alignment. The tree produced is unrooted. In the following alignment of 4 sequences, only variable sites are shown: Sequence Sequence Sequence Sequence 1 2 3 4 1 1 1 1 1 1 1 1 1 1 2 2 1 2 3 4 5 6 7 8 9 0 1 2 3 4 5 6 7 8 9 0 1 C.C.T.A.T.G.T.C.T.C.A.T.G.C.G.C.G.T.C.C.G C.G.C.A.T.A.T.T.T.C.G.T.G.A.G.T.G.A.T.T.A T.G.T.A.C.G.C.C.C.T.G.C.C.A.A.C.A.T.T.T.G C.G.C.G.T.G.T.T.T.C.G.T.C.A.G.T.C.T.C.T.A 1) Which is the maximum parsimony tree for these four sequences? [Count the number of informative sites supporting each of the 3 possible unrooted topologies. In a tree with 4 sequences, the topology supported by the largest number of sites must be the maximum parsimony topology.] 2) How many nucleotide substitutions are inferred to have occurred on each branch of the MP tree estimated in (1) above? [In a maximum parsimony analysis, some sites in the sequence alignment provide ambiguous information concerning the positions of substitutions on the tree. Nevertheless, a probabilistic approach can be taken: for informative sites in conflict with the most parsimonious tree, 1/2 of a nucleotide substitution can be assigned to each of the terminal branches. This simple solution is true only for 4 taxon trees.] Terminal Branch to sequence 1: substitutions Terminal Branch to sequence 2: substitutions Terminal Branch to sequence 3: substitutions Terminal Branch to sequence 4: substitutions Central Branch: substitutions 2 3) When alternative types of nucleotide substitution do not occur at equal frequencies, it may be better to perform “weighted maximum parsimony”. For example in mammalian mitochondrial DNA, transitions are thought to occur (at least) 10 times as often as transversions. Therefore, to weight we could count each transversional change as worth 10 transitions. If we do this, which is the maximum parsimony tree? 3 Part II: Distances and UPGMA Trees The objective of this exercise is to calculate distances and build a UPGMA tree for the following group of sequences: 1 Seq1 Seq2 Seq3 Seq4 Seq5 Seq6 50 GATTGCCTTG .......... AG........ ...C...... ...G...... ..CC...... GATCCTTGAC .......... .......... A.C....... A.C....... ..G.....G. TGATCGGGTC .....A.... .....A.... .A....A... .A....A... ......AC.. GCTTGCTACC .......... .......... .......... .......... ...C...G.. TTACAGCGAT .......... .G..G...G. .G..T..... .G..C..A.. .C..G..C.. 1) Count the number of differences Seq1 Seq2 Seq3 Seq4 Seq5 Seq6 Seq1 - Seq2 Seq3 Seq4 Seq5 Seq6 - 2) Calculate the uncorrected p distances Seq1 Seq2 Seq3 Seq4 Seq5 Seq6 Seq1 - Seq2 Seq3 Seq4 Seq5 Seq6 - 3) Correct the distances to account for multiple hits using the Jukes Cantor (JC) model. Kxy = - 3/4 ln (1 – 4/3 d) Where: Kxy = estimated number of nucleotide substitutions per site between sequence x and sequence y [estimated distance between sequence x and sequence y] d = observed proportion of nucleotides that differ between the 2 sequences Seq1 Seq2 Seq3 Seq4 Seq5 Seq6 Seq1 - Seq2 Seq3 Seq4 Seq5 Seq6 - 4 4) The distances below were estimated using the F84 model. This model is very similar to the HKY85 model and allows transitions and transversions to have different rates as well as allowing for unequal base frequencies. Seq1 Seq2 Seq3 Seq4 Seq5 Seq6 Seq1 Seq2 Seq3 Seq4 Seq5 0.0202 0.1311 0.1558 0.1824 0.2643 0.1075 0.1798 0.2073 0.2914 0.2286 0.2578 0.3589 0.0622 0.2705 0.3043 Seq6 How do the distances estimated using the F84 model differ from those estimated using the JC model? Which model do you think is more appropriate for the sequence data and why? 5) Draw the UPGMA tree for the 6 sequences based on the distance matrix in #4. 5 6) The neighbour-joining tree for these 6 sequences is given below: How do the UPGMA and NJ trees differ? 7) Based on observations of the sequence data and your knowledge of the assumptions of UPGMA and NJ, which tree do you think is most accurate and why? 8) It is known that sequences 1 and 2 diverged 2 million years ago. Calculate the rate of substitution using the distances in (3). r = K/2T where: r = rate of substitution (number of substitutions per site per year) K = number of substitutions between 2 sequences T = time since divergence of the 2 sequences 6 9) Assuming a molecular clock, calculate the age of : a) the divergence of sequences 1+2 from 3 b) the divergence of sequences 1+2 from 4+5 (excluding 3) 10) In a relative rates test, we can compare the rates in two closely related species (or clades of species), A and B, with a third more distantly related outgroup species (or clade of species), C, as a reference. KAC = average distance between A and the outgroup taxon C KBC = average distance between B and the outgroup taxon C If KAC-KBC = 0 then both lineages are evolving at the same rate. Anything other than 0, then one lineage is evolving faster than the other. Conduct a relative rates test to determine whether: a) sequence 3 is evolving at the same rate as sequences 1 and 2 b) sequences 4 and 5 are evolving at the same rate as sequences 1 and 2 11) Is it appropriate to use a molecular clock to estimate the time of a) the divergence of sequences 1+2 from 3? b) the divergence of sequences 1+2 from 4+5 (excluding 3)? 7 Part III. Bacterial Phylogenies a) Comparing Tree Topologies Eight evolutionary trees are shown overleaf. The same eight species of bacteria appear in each, but each tree was derived from analysis of a different protein. Species Gene Length Protein Escherichia coli Haemophilus influenzae Neisseria meningitidis Pasteurella multocida Pseudomonas aeruginosa Vibrio cholerae Xylella fastidiosa Yersinia pestis rpoA recA rpsA ffh mopA rplB tpiA tufA 327 343 545 450 541 271 243 394 DNA- directed RNA polymerase alpha subunit RecA recombination protein 30S ribosomal subunit protein S1 signal recognition particle protein chaperonin 60kD subunit (GroEL) 50S ribosomal subunit protein L2 triosephosphate isomerase elongation factor Tu Neisseria meningitidis is placed in the beta proteobacteria; the other seven species are classified as gamma proteobacteria. The length given is the number of aligned amino acid sequences. Tree methods used: after multiple sequence alignment, a matrix of pairwise distances was constructed; distances were corrected for multiple hits by Kimura’s empirical method. The trees were then estimated by Neighbour-joining (NJ) analysis. NJ defines the tree topology and branch lengths but produces an unrooted tree. 1) In the rpoA tree, which species is/are the most closely related to Vibrio cholerae? 2) The majority of the eight trees have identical topologies. Which trees have different topologies, and how do they differ from the consensus topology? 3) What are the possible reasons for differences in topology among these trees? 4) How might you root the tree? Which of the trees looks like the best representation of a rooted tree for these species? 8 rpoA recA mopA rplB rpsA tpiA ffh tufA 9 b) Neighbour Joining Analysis with Bootstraps The sequence of the infC gene encoding translation initiation factor 3 is available for 15 different species of gamma proteobacteria. These predicted protein sequences together with that from Neisseria meningitidis were analysed by the NJ method as on the previous pages. The infC gene is very unusual in that analysis of the E. coli gene has revealed that its translation initiation codon is ATT. In other bacterial species, other unusual codons are used: the inferred translation initiation codons for each species are given here: 1. Buchnera sp. APS 2. Buchnera aphidicola 3. Escherichia coli 4. Haemophilus influenzae 5. Klebsiella pneumoniae 6. Neisseria meningitidis 7. Pasteurella multocida 8. Proteus vulgaris ATT ATT ATT ATT ATT ATC ATT ATT 9. Pseudomonas aeruginosa 10. Pseudomonas fluorescens 11. Pseudomonas syringae 12. Salmonella typhimurium 13. Serratia marcescens 14. Vibrio cholerae 15. Xylella fastidiosa 16. Yersinia pestis ATC ATT ATT ATT ATT ATT ATC ATT Neisseria meningitidis is placed in the beta proteobacteria; the other fifteen species are classified as gamma proteobacteria. The results of the NJ analysis are summarised below. The NJ method progresses through n-2 rounds (where n is the number of sequences), inserting a new internal branch in each round until a fully bifurcating tree is produced. Each time a new branch is inserted it implies that 2 sequences (or 2 clades of sequences) have been joined at one end of the branch. The notation below is output from the NJ tree construction program NEIGHBOR of the PHYLIP package. It indicates which sequences (or clades) are joined in each round, and shows the branch length from that node to the sequence (or clade). In this notation, clades are referred to as ‘nodes’ on the tree and are given a number referring to the lowest numbered sequence within that clade. round round round round round round round round round round round round round round 1 : 2 : 3 : 4 : 5 : 6 : 7 : 8 : 9 : 10: 11: 12: 13: 14: seq 10 (0.008) joins seq 11 (0.010) seq 9 (0.063) joins node 10 (0.054) seq 6 (0.197) joins seq 15 (0.361) seq 4 (0.025) joins seq 7 (0.030) node 6 (0.092) joins node 9 (0.181) seq 1 (0.050) joins seq 2 (0.058) node 4 (0.132) joins node 6 (0.056) node 4 (0.024) joins seq 14 (0.160) seq 13 (0.035) joins seq 16 (0.021) seq 3 (0.003) joins seq 12 (0.003) node 3 (0.006) joins seq 5 (0.018) seq 8 (0.074) joins node 13 (0.035) node 1 (0.063) joins node 3 (0.037) node 1 (0.006) + node 4 (0.054) + node 8 (0.009) 10 For example, in round 1, sequence 10 (see above) joins sequence 11 at the first node. The branch length from this node to sequence 10 is 0.008 and to sequence 11 is 0.010. In round 2, sequence 9 joins node 10 (i.e., the clade formed by sequences 10 and 11) at a second node. The branch length from this second node to sequence 9 is 0.063 and from the second node to the first node is 0.054. The output can also be represented in the form of a treefile: ((((((10:0.008,11:0.010):0.054,9:0.063):0.181,(6:0.0197,15:0.361):0.092):0.056,(4:0.0 25,7:0.030):0.132):0.024,14:0.160):0.054,((1:0.050,2:0.058):0.063,((3:0.003,12:0.003 ):0.006,5:0.018):0.037):0.06,((13:0.035,16:0.021):0.035,8:0.074):0.009); 1) Draw out the full tree [Do this roughly at first, concentrating on the topology only. Then draw out a neater version with branch lengths roughly to scale. Remember that the tree is unrooted! 11 2) Re-draw the tree as a rooted tree by rooting on an appropriate outgroup taxon. 3) An alternative approach to rooting a tree is to assume an approximate molecular clock, and then root at the midpoint of the longest pairwise distance implied by the tree. In this tree, the longest pairwise distance is from Buchnera aphidicola to Cylella fastidiosa. The distance is given by the sum of all the branches connecting these species (0.058 + 0.063 + 0.006 + 0.054 + 0.024 + 0.056 + 0.092 + 0.361 = 0.714). With midpoint rooting, on which branch of the tree does the root lie? How does this compare with the outgroup rooting above? 12 4) One thousand bootstrap replicates were examined to assess the support for the infC tree. The matrix below shows the results. Each column refers to a sequence. Each row refers to a partition in the tree (i.e., a branch separating 2 clades) between the sequences indicated by * in one clade and the sequences indicated by – in the other clade. The value at the right on each row is the number of bootstrap replicates in which the partition was found. 1234 **** **** **** ******* --** ********** **-* **-* **** ---* 5678 **** **** *-** **-* *-** **** *--* *--* **** **** -*** ***-*** 111 9012 *--* ---* **** **** ---* **** ---* ---* **** ********** ***- 1111 3456 **** **** **-* **** **-* **** **-* *--* -****** **** -****** 1000 1000 945 999 845 1000 654 991 979 709 998 443 393 Place the bootstrap values on the tree in (2) 5) Is the tree topology obtained for infC consistent with any of those found on page 8? If yes, which? If no, how does it differ? 6) What is the minimum number of times that the nature of the translation initiation codon must have changed on the infC NJ tree? 7) Do the three Pseudomonas species form a monophyletic group in the infC NJ tree? If so, can this clade be considered to be well-supported or poorly supported in the analyses of the infC dataset? 8) Do Vibrio, Haemophilus and Pasteurella form a monophyletic group in the infC NJ tree? If so, can this clade be considered to be well-supported or poorly-supported in analyses of the infC dataset? 9) Is the grouping of Escherichia/Salmonella/Klebsiella with the Buchnera species in the infC NJ tree reliable?









