File - Serrano High School AP Biology
advertisement

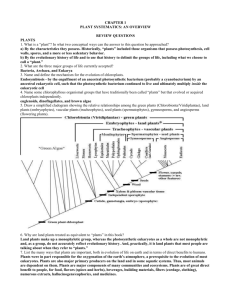
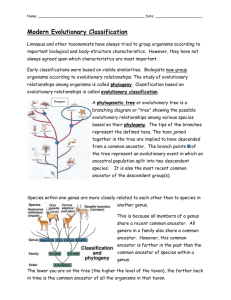
Classification: Phylogeny and Systematics The fossil record reveals that the diversity of organisms that have populated the earth is the result of three broad paths of speciation, with the effects of extinction. In phylogeny, our goal is to trace the evolutionary history of each species. Reconstructing the phylogeny is part of systematics, where you also have to classify species based on their history. 1) Phyletic Change: Adagenesis Under pressures of directional selection-- a species gradually accumulates changes until it is so distinct from the original population that it is a new species. A B (You’ll need to review all the types of selection. Each arrow represents time and natural selection.) 2) Cladogenesis: Splitting of lineages. Species formed by cladogenesis are descendants of a common ancestor. This can occur radially instead of linearly. A A B 3) Adaptive Radiation: Sudden diversification of a group of organisms that share a common ancestor that itself has newly evolved. This is a combination of cladogenesis and phyletic change. Also occurs radially. CABD | | F E From these types of speciation, we can create a phylogenetic tree. Phylogenetic trees (Evolution Trees) show the following: 1) Common ancestry 2) Relative divergence from the ancestor 3) Evolutionary relationships in reference to living representatives of the groups. Phylogentic trees start off with a common ancestor. Speciation occurs in the common ancestor population with changes in the gene pool. New populations (new species) branch off from the common ancestor population. These new populations will be exposed to environmental conditions, and, over time these populations will undergo evolution and speciation, thus creating new branches and populations. Populations with a recent common ancestor are closely related evolutionarily and the branches should be close to each other. The organisms in these closely related groups would tend to exhibit homologous structures (similar structures, may have different functions), similar DNA sequences (and similar biochemistry), and similar embryonic development. The development of new species from a common ancestor is not necessarily a linear development. It is more of a radial event that shows more than one species developing from a common ancestor. Two evolution theories of species development: 1 1) Gradualism: change occurs at a regular rate, gradually over time. 2) Punctuated Equilibrium: refers to the tempo of evolution, assuming that new species formed principally in small populations on geographic periphery of the range of the species, speciation occurred rapidly, and the new species then outcompeted the old ones. The population changed faster at sometimes than at other times. These changes can come about by sudden changes in environmental conditions. Nils Eldredge and Stephen J. Gould developed this theory. Classification (evolutionary history) Scheme: Categories and Evolutionary Theories We have to take into account variability, relationships, and evolutionary relatedness to classify organisms into various categories. The largest categories are called Kingdoms (for Linneaus) and the smallest are Species. (Note: Currently, there is a three-domain system suggested. Domains would then be the largest category.) Binomial, meaning two names refers to the way scientists label living things. The two names come from the genus and species of the classification system. This system was developed by Karl Von Linne, who latinized his own name to Carl Linneus (17011778). He is responsible for creating categories within categories. He chose to use Latin because of two reasons: 1) Latin is not commonly spoken anywhere outside of a few academic halls or in religious ceremonies and not likely to change 2) Latin forms the roots of many words in a number of languages. In a monumental feat, Linneus single-handedly named and classified many of the earth's organisms. Since Linneus' time, the rules for assigning names are rigidly enforced by international commissions. Here are the categories for classifying organisms from the highest to the lowest level. Categories = How to remember them Domain = Did or Kingdom = King or Phylum = Philip or Class = Come or Order = Over or Family = From or Genus = Greece or Species = Singing or Does King Philip Come Over For Great Salad These are just names applied to groups of organisms that are thought to be related to one other. A taxon is the taxonomic unit at every level. For example, Mammalia is the taxon at the class level. For each taxon, there can be a super category (above the taxon) and a sub category (below the taxon). E.g. …..super phylum, phylum, subphylum…. Taxonomy has two main objectives: 1) To sort out closely related organisms and assign them to separate species and to name newly discovered species. 2) To organize species into the higher taxonomic categories. 2 Phylogeny: evolutionary history of a species or a group of related species. Systematics: the science of classification with the use of taxonomies. Biologists wanted taxonomies to be not only convenient and useful but also to accurately reflect the phylogenetic relationships among organisms. These taxonomies are hypotheses about evolutionary histories. Some of these hypotheses can be tested and revised if necessary. The study of evolutionary relationships is known as EVOLUTIONARY SYSTEMATICS. The modern phylogenetic systematics is based on cladistics. The modern diagrams are called cladograms— phylogenetic trees. The tree is constructed from a series of dichotomies (branches). Each branch represents a divergence of two species from a common ancestor. The sequence of branching symbolizes when speciation took place—historical reference. An evolutionary branch of the tree is called a clade. A clade is a true clade when it contains the ancestral species and all of its descendents—called monophyletic. A classification scheme that accurately reflects evolutionary history is termed MONOPHYLETIC. This means that at each level, all members of the taxon should be the descendants of a common ancestor. Monophyletic classification is difficult to achieve for two reasons. 1) It is difficult to know completely the evolutionary histories of organisms. 2) Convenience and utility may be more important than an accurate reflection of phylogeny. Polyphyletic Taxon: We usually see that a taxon contains members that have descended from more than one common ancestor. Branches don’t share a common ancestor. Paraphyletic Taxon: excludes species that share a common ancestor that gave rise to the species included in the taxa. A branch that excludes other species form the common ancestor. The goal of cladistics is to construct a HOLOPHYLETIC TAXA. The holophyletic taxa must contain ALL of the descendants from the common ancestor plus the ancestor. Schools of Taxonomy: Two alternative methodologies: 1) Phenetics 2) Cladistics These are supposedly objective taxonomic methods. 1) Phenetics: This is based only on a species' observable characteristics. These characteristics are divided into 'unit characteristics.' These unit characteristics are assigned a value, and a computer compares these values. Homology/Analogy is not considered. Phenetics suggests that the problems with this method are resolved if enough characteristics are compared. 2) Cladistics: This is based on phylogeny. Cladistics suggests that the branching of one lineage from another in the course of evolution is the one event that can be determined objectively. This method considers only homologies when constructing the cladogram. 3 Taxonomic Methods: Several steps for the traditional taxonomic method: 1) An organism is assigned a taxon based on the outward similarities to other organisms in the taxon. 2) The similarities are tested for homologies. Fossil evidence is examined whenever possible. 3) Compare DNA/RNA/Proteins of organisms, whenever possible. 4) Various stages in the lifecycle and the patterns of embryonic development are also compared. When constructing a taxon in the traditional way, two factors need to be considered. 1) Some factors produce evidence of genealogy (branching patterns that show the evolutionary history of the organism). 2) The taxon should reflect the degree to which the organisms have diverged since they began to travel in different evolutionary paths. Traditional taxons contain information about the sequence in which branching occurred and the extent of the biological changes. Problem: Not all similarities are due to a common ancestor; some are due to convergent evolution. However, the more complex two similar structures, the more improbable it is that they came from two separate ancestors. Tools of Taxonomists: 1) Homology and Phylogeny: Systematics is largely a comparative science. A systematist classifies, to a large extent, on similarities or morphology (homology/analogy). Organisms are grouped in a process based on similarities in structure and other phenotypic characteristics. Superficial similarities are not always useful in deciding the taxon for the organism. A big question in evolutionary systematics concerns the origin of similarities or differences between two organisms. Does the similarity reflect inheritance from a common ancestor, or does it reflect an adaptation to similar environments by organisms that do not share a common ancestor? Does a difference between two organisms represent different phylogenetic histories or does it reflect the adaptation of closely related organisms to different environments? Similarities that have a common origin but do not necessarily the same function are termed HOMOLOGOUS. Structures that have a similar function, but have different evolutionary backgrounds are said to be ANALOGOUS. Modern systematics relies on both morphology and DNA/RNA/Protein comparisons. 2) Molecular Biology. A) Amino Acid Sequences: Compare the amino acid sequences of the same protein in different organisms. The greater the number of amino acid sequence differences, the more distant their evolutionary relationship. The smaller the amount of differences, the closer the relationship. Some biologists suggest that a change in an amino acid represents a functional difference in the 4 molecule. Other biologists say that amino acid changes occur at random; and that they do not represent the result of the selection process, such changes simply mark the passage of time. Instead of functional differences, changes in amino acid sequences are a MOLECULAR CLOCK. This clock can be used to determine the time at which groups diverged. B) Nucleotide Sequences/DNA Observations: Since it is now possible to sequence nucleotides, the comparison of homologous proteins has been largely abandoned. Nucleotide sequencing is more sensitive and easier to do than comparing amino acids. The nucleotide sequences of different species are placed into a computer and compared. Taxonomic relationships can be determined with these comparisons. DNA-DNA Hybridization: This is used to compare the DNA strands of different species. It is an important tool in conjunction with other methods. Restriction Mapping: A process, which obtains restriction fragments (RFLP) with restriction endonucleases and compares this with the restriction fragments of other species. The mitochondrial DNA (mtDNA) of several species can be compared in this way. You can use PCR with this tool. C) Molecular Clocks: Proteins evolve at different rates, but for a given protein the rate seems to be pretty constant. If homologous proteins are examined and compared, the number of amino acid differences is proportional to the amount of time that has elapsed since the branching of the populations. DNA comparisons are more promising than protein comparisons. Molecular clocks are calibrated by graphing the number of amino acid and/or DNA base pair differences against time. This is done for a series of evolutionary branch points from the known fossil record. If no clear fossil evidence is present, the graph then estimates the time of species divergence. The consistent rate of protein and DNA changes implies a large number of neutral mutations. This is a controversial point among scientists who use the molecular clock. Back to the diversity of organisms: In the beginning of taxonomy (naming things) there were two kingdoms; either something was an animal or a plant. From there scientists devised a three-kingdom system-- plants, animals and protists. Currently, scientists use a five-kingdom system. Currently a three-domain system is being suggested. The domains are Eukarya, Archae, and Bacteria. Each domain is based on a cell type: Eukarya—eukaryotes, Archae – archaebacteria, and Bacteria – eubacteria. Interestingly, Eukarya and Archae shared a common ancestor, where as, bacteria branched off earlier. Kingdoms: Moneran: Prokaryotes: bacteria and cyanobacteria 5 Protista: Eukaryotic: various simple, single celled eukaryotes including protozoa (formerly animals) and Chlorophyta (formerly plants). In the three-domain system, there are many more kingdoms where the protist kingdom once stood. We’ll call Protista a Super Kingdom with Kingdoms branching off of the super kingdom. Fungi: Eukaryotic: non-photosynthetic organisms. Plants: Eukaryotic: non-vascular and vascular plants. Animals: Eukaryotic: multicellular from sponges to humans. Note: viruses are not included in this scheme. 6