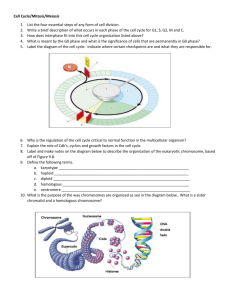
Total number of F2 offspring
advertisement

2.1 Variation and Mutation Sexual reproduction produces offspring which are genetically different from both parents and all other members of the species. Sexual reproduction maintains and increases genetic variation within a population. Chromosomes are thread-like structures carrying genetic code. They are found in the nucleus of a living cell. A chromatid is one of two identical replicas of a chromosome. Homologous chromosomes match each other gene for gene. A diploid cell has a double set of chromosomes (2n). A haploid cell has a single set of chromosomes (n). Meiosis is a form of cell division which results in the production of four haploid (n) gametes from one diploid (2n) mother cell. Gamete mother cells produce cells which will undergo meiosis. Gamete mother cells can be found in the testes and ovaries of animals and anthers and ovaries of flowering plants. Meiosis involves two nuclear divisions. During the first meiotic division the cell becomes two haploid cells. During the second meiotic division four different haploid cells (gametes) are formed. During division 1 homologous chromosomes line up at the equator. Their final position is random relative to any other pair. This leads to random assortment during subsequent meiotic divisions. Random assortment brings about different genetic combinations and may therefore result in formation of new phenoypes in the next generation. During division 1 homologous chromosomes pair up. Members of the pair are joined at points called chiasmata. Crossing over can occur at chaismata. This is when two chromatids swap portions of genetic material by breaking and joining the end of the other chromatid. Crossing over also results in new genetic combinations and results in the formation of new phenotypes in the next generation. Both crossing over and independent assortment lead to increased variation within a species by producing new combitions of genetic material in gametes. In the long term such variation is important as it helps species adapt to a changing environment. A cross between two true breeding parents which possess different forms (alleles) of TWO genes is called a dihybrid cross. The expected phenotypic ratio of the offspring of a dihybrid cross is 9:3:3:1 If the two alleles of two different genes are located on the same chromosome (and hence move together during meiosis) they are said to be linked. Linked genes can be separated during crossing over. Genes which are closer together on a chromosome have less chance of being separated and two with greater distance between each other. The distance between a pair of linked genes is indicated by the percentage number of F2 recombinants which result from a test cross. The recombination frequency or crossover value (COV) is calculated as shown: COV = Number of F2 recombinants x 100 Total number of F2 offspring COV of 1% represents a distance of one unit on a chromosome. The position of a gene on a chromosome is called its locus (loci) The separation of linked genes is another source of variation within a species. Genes found on the X chromosome which are not present on the smaller Y chromosome are said to be sex-linked. Examples of conditions brought about by sex linked genes include colour blindness and haemophilia. Sex chromosomes must be represented by symbols X and Y and alleles in appropriate upper case and lower case superscripts eg. XªXª or XªY. A mutation is a change in the structure of mass of an organisms genetic material. Non-disjunction occurs if spindle fibres fail during meiosis and members of a pair of homologous chromosomes fail to separate. This type of mutation changes the number of chromosomes in a cell. Non-disjunction of chromosome pair 21 in a human egg cell leads to Down’s Syndrome. Complete non-disjunction is when all spindle fibres fail, and all homologous pairs fail to separate. This results in diploid gametes. Fertilisation involving such gametes produces mutant plants with an extra set of chromomosomes. This type of chromosome mutation is called polyploidy. Polyploid plants are normally larger, may produce larger seeds, fruits and therefore can be of economic importance. Chromosome mutations involve a change in the number or sequence of genes in a chromosome. There are four different way in which this can happen: o Deletion – when a gene or genes are lost from the chromosome o Duplication – when genes from the chromosome’s homologous partner become inserted somewhere along its length. o Translocation – when a section of one chromosome breaks and attaches to another chromosome which is not its homologous partner. o Inversion – when a section breaks from a chromosome, turns round before joining up again on the chromomsome, leading to a reversal of the normal sequence in the affected section. Gene mutations involve a change in or one or more of the nucleotides in a strand of DNA. There are four different way in which this can happen: o Substitution – when one base is replaced by another o Inversion – when bases on a strand swap places o Deletion – when a base or bases are lost from the DNA strand o Insertion - when a base or bases are inserted into the DNA strand Substitution and Inversion are called point mutations. They usually bring about only minor changes and the organism is usually only slightly affected or not at all. Deletion and Insertion are known as frameshift mutations. These lead to major changes as large portions of the genes DNA is misread. Mutation rate can be increased artificially by chemical agents or irradiation (mutagenic agents such as mustard gas, X-rays, UV light, gamma rays)