Supplementary Data (doc 792K)
advertisement

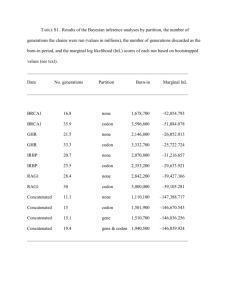
PIKE-OVERZET et al PRECLINICAL LENTIVIRAL RAG1 GENE THERAPY Supplemental Data Supplemental Materials and methods Vector construction and lentiviral production Lentiviral gene transfer constructs were derived from pRRL.PPT.PGK.GFPpre(1). The pRRL.PPT.EFS.RAG1.pre (hereafter: EFS.RAG1), pRRL.PPT.SFFV.RAG1.pre (hereafter: SFFV.RAG1), pRRL.PPT.EFS.coRAG1.pre (hereafter: EFS.coRAG1), pRRL.PPT.SFFV.coRAG1.pre (hereafter: SFFV.coRAG1) and pRRL.PPT.UCOE.coRAG1.pre (hereafter: UCOE.coRAG1) transfer vectors were constructed by replacing the PGK promoter by a human EFS ( S for short form) promoter, an enhancer–promoter from the spleen focus-forming virus (SFFV) or a ubiquitously acting chromatin opening element (A2UCOE). The GFP sequence was replaced by native human RAG1 cDNA or the codon-optimized version of RAG1 (coRAG1). Codon optimization of the human RAG1 gene was performed by Geneart (Regensburg, Germany). The WPRE used in these constructs is lacking the X protein ORF and 4 possible ATGs were mutated(2). Gene transfer constructs were validated by full DNA sequencing of the transgene. For Supplemental figures 2 and 3 the gene transfer vector was constructed as follows: The backbone of replication-defective, self-inactivating Trip-ΔU3-EF1α-RAG1 lentiviral vector has been previously described.(3, 4) The RAG1-containing construct were generated using BamH1 (New England Biolabs, Ozyme, Saint Quentin Yveline, France) ligation of the coding sequence of native human RAG1 cDNA. Large-scale helper-plasmid preparations were produced by PlasmidFactory (Bielefeld, Germany). 293T cells were transiently transfected with transfer and helper plasmids using the calcium-phosphate method or FuGENE 6 transfection reagent (Roche, Basel, Switserland). Lentivirus was harvested 24h, 32h and 48h after transfection. Cleared supernatant was filtered through 0.22 µm pore cellulose acetate filters and stored at – 80°C. Pooled lentiviral supernatant was concentrated by ultracentrifugation for 16 hours at 18,000 x g and 4°C. Pellets were resuspended in StemSpan serum-free expansion medium (StemSpan-SFEM, Stemcell Technologies Inc, Vancouver, BC, Canada). Because suitable antiRAG1 antibodies were not available, we determined the viral titer using RQ-PCR as described below. RQ-PCR Total RNA from viral supernatant and cells was extracted using the RNeasy Total RNA isolation kit (Qiagen, Hilden, Germany). RNA was reverse transcribed into cDNA. Genomic DNA was extracted using the GeneElute Mammalian Genomic DNA miniprep kit (Sigma-Aldrich, Saint Louis, MO, USA). RQ-PCR reactions were performed on the ABI Prism 7700 Sequence Detector System (Applied Biosystems, Foster City, CA). The following primers and probes were used. WPRE: forward primer, 5’-CCGTTGTCCGTCAACGTG-3’; reverse primer, 5’AGTTGACAGGTGGTGGCAAT-3’; probe, 5’-FAM-TGCTGACGCAACCCCCACTGGCTAMRA-3’. RAG1: forward primer, 5’- GCCAAACCTAACTCTGAACTGTGTT-3’; reverse primer, 5’- GCGTCTCGTGGTCAGACTCA-3’; probe, 5’-FAM-CCATTGTGCCTTATGCTTAMRA-3’. coRAG1: forward primer, 5’-CAACTGCAAGCACGTGTTCTG-3’; reverse primer, 5’-GCAGTAGCTGCCCATCACTTT-3’; probe, 5’-FAMAGAGTGTGCATCCTGCGGTGCCT-TAMRA-3’. For copy number quantification of unknown samples, standard curves were created by preparing ten-fold serial dilutions of plasmid constructs of known concentration that contained the WPRE sequence. For samples derived from cellular RNA, the expression levels were normalized to the expression of the ABL gene: forward primer, 5’-TGGAGATAACACTCTAAGCATAACTAAAGGT-3’; reverse primer, 5’GATGTAGTTGCTTGGGACCCA-3’; probe, 5’-FAMCCATTTTTGGTTTGGGCTTCACACCATT-TAMRA-3’. 1 PIKE-OVERZET et al PRECLINICAL LENTIVIRAL RAG1 GENE THERAPY Determining virus titers of non-EGFP vectors Using a lentiviral RAG2.IRES.EGFP construct, we found that viral titers as determined by RQPCR were approximately 65 times higher (most likely due to detection of intermediates of viral replication) than transduction-based viral titers (data not shown), allowing us to make a rough estimation of the multiplicity of infection (MOI) of the non-EGFP vectors. Flow cytometry studies and cell sorting Peripheral blood samples were taken periodically from live mice and peripheral blood, spleen, thymus and bone marrow were obtained from CO2-euthanized mice and cell suspensions were prepared. Cells were counted and their surface markers stained with the following anti-mouse antibodies in various combinations: fluorescein isothiocyanate (FITC)-conjugated IgD (1126c.2a) and T-cell receptor β (TCRβ) (H57-597); R-phycoerythrin (PE)-conjugated CD19 (ID3), IgM (R6-60.2) and TCR γδ (GL3); peridinin chlorophyll protein (PerCP)-conjugated CD8α (536.7); allophycocyanin (APC)-conjugated CD3ε (145-2C11) and CD19 (ID3); PE-Cy7-conjugated CD4 (L3T4) and CD45R/B220 (RA-6B2); biotin-conjugated CD11b (M1/70), CD43 (S7), CD45R/B220 (RA-6B2), GR-1 (RB6-8C5), NK1.1 (PK136) and TER-119 (Ly-76) (all antibodies BD Biosciences, Santa Clara, CA, USA). When necessary, streptavidin-conjugated APC-Cy7 (BD Biosciences) was added as a second step. Data were acquired on a FACSAria or a FACSCanto II (BD Biosciences). Analyses were performed using FlowJo software (Treestar, Ashland, OR, USA). CD19+ B cells and CD11b+ cells were sorted from pooled SFFV.RAG1 spleen cells using a FACSDiva (BD Biosciences). Immunizations and details ELISA A sandwich enzyme-linked immunosorbent assay (ELISA) was used to determine IgM, IgG1 or IgG serum levels using unlabeled and peroxidase-labeled anti-mouse IgM, IgG1 and IgG antibodies (SouthernBiotech, Birmingham, AL, USA) for capture and detection, respectively. Serially diluted sera were incubated at room temperature for 3 h, and azino-bisethylbenzthiazoline sulfonic acid (ABTS) was used as a substrate. For TNP-specific enzymelinked immunosorbent assay, plates were coated with TNP–KLH and serially diluted pre- and post-immunization sera were incubated for 2 h. For detection, biotinylated anti-IgG and streptavidin-coupled peroxidase were used, followed by ABTS as substrate. Antibody concentrations were calculated by using purified IgM, IgG1 and IgG proteins as standards. Supplemental Results Two months after transplantation into Rag1-/- mice, T lymphocytes were found in the peripheral blood and in the peripheral organs (supplemental figure 2) of 5 out of 30 mice (17%) (supplemental Table 1). Despite low T-cell counts in all the organs, the cells were functional (Data not shown; in vitro proliferation assays n=1). Thymic reconstitution was low (4-6% CD4+CD8+ cells). Of note, we did not observed any reconstitution of the B-cell compartment. hRAG1 VCNs in central and peripheral lymphoid organs was higher in the reconstituted mice as compared to the non-reconstituted mice (supplemental figure 3). We observed higher VCNs and hRAG1 expression in the mice who had the higher T-cell counts (VCN= 8 in the spleen, VCN= 2 in the BM and VCN=16 in LN, VCN=6 in the SCA1 cell before transduction). Altogether, these data led us to conclude that RAG1 transgene expression was not sufficient to restore the T- and B-cell development using this particular construct. 2 PIKE-OVERZET et al PRECLINICAL LENTIVIRAL RAG1 GENE THERAPY Supplemental Table 1. Overview of B and T cell reconstitution in RAG1-/- mice transplanted with EF-1a-RAG1 transduced Sca-1 cells. Irradiation dose 3 Gy 8 Gy 8 Gy Cells injected Sca-1 Sca-1 Sca-1 Sca-1 Sca-1 Sca-1 Sca-1 Sca-1 CD4+/CD3 +% 17 12 7 29 5 20 18 15.7 11.8 ± 5.8 PBL CD8+/CD3 B220+/IgM +% +% 1.6 <1 5.4 <1 3 1.6 20 35 0.3 <1 14 42 6 <1 12.4 47 3.2 ± 2.4 <1 Test PBL (months) 4 5 5 5 5 Thymus CD4+/CD8+% (1*106 cells) (2*106 cells) (800 000 cells) (22*106 cells) (140 000 cells) (2.8*106 cells) Not done 85 (10*106 cells) 4 5 6 76 4 45 Supplemental Table 2. Overview of B and T cell reconstitution in Rag1-/- mice in figure 2 A-D Reconstitution Transplanted cells WT Rag1-/EFS.coRAG1 SFFV.coRAG1 # of mice 7 5 9 7 Success rate B cells T cells B cells T cells 7/7 0/5 5/9 6/7 7/7 0/5 5/9 6/7 100% 0% 56% 86% 100% 0% 56% 86% Supplemental Table 3. Overview of B and T cell reconstitution in Rag1-/- mice in figure 2 E, F and figure 4 Reconstitution Transplanted cells WT Rag1-/UCOE.coRAG1 SFFV.coRAG1 # of mice 4 2 6 6 Success rate B cells T cells B cells T cells 4/4 0/2 6/6 6/6 4/4 0/2 4/6 4/6 100% 0% 100% 100% 100% 0% 66.6% 66.6% 3 Reconstitution 3/18 Control mouse 1/3 Control mouse 1/9 Control mouse 5/30 (17%) PIKE-OVERZET et al PRECLINICAL LENTIVIRAL RAG1 GENE THERAPY Supplemental Figure legends Supplemental figure 1. Partial complementation of the Rag1-/- phenotype by tranduced lineage negative cells. (a) Flow cytometry analysis of surface IgD/B220 expression in peripheral blood 6 weeks after transplantation. The numbers in the graphs represent the percentage of B220+IgD+ cells of MNCs. (b) Average B-cell percentage of lymphocytes 14 weeks after transplantation, determined by flow cytometery. * P value = 0.0022. Statistical significance was determined by a two-tailed, unpaired t test. (c) Flow cytometry analysis of surface CD4 and CD8 expression of thymocytes. The numbers in the graphs represent the percentage of total thymocytes of each quadrant. (d) Genescan analysis of PCR products obtained from amplifications using Cµ or Cγ-specific primers in combination with a consensus VH primer. Detection of polyclonal rearrangements on cDNA derived from spleen cells, 14 weeks after transplantation of transduced lin- cells. (e) 14 weeks after transplantation, IgM and IgG1 concentrations in the serum of mice that received WT, Rag1-/-, EFS.RAG1 or SFFV.RAG1 transduced lin- cells were determined by ELISA. Supplemental figure 2. Partial complementation of the Rag1-/- phenotype by tranduced Sca-1+ cells. Two months after transplantation, peripheral blood was analyzed for the presence of CD4+/CD3+ and CD8+/CD3+ T cells as well as B220+ B cells. (a) T- and B-cell concentrations in peripheral blood. (b) Absolute numbers of cells of each population in peripheral lymphoid organs. Absolute numbers were determined based on the total cell count of each organ and the relative size of the populations as determined by flow cytometry. TG: Gene therapy, ctrl: control Supplemental figure 3. Vector copy numbers. Two months after transplantation, VCNs were determined for central and peripheral lymphoid organs in reconstituted and non-reconstituted mice. 4 PIKE-OVERZET et al PRECLINICAL LENTIVIRAL RAG1 GENE THERAPY Supplemental Figures Supplemental Figure 1 54,08 Supplemental Figure 2 5 PIKE-OVERZET et al PRECLINICAL LENTIVIRAL RAG1 GENE THERAPY 6 PIKE-OVERZET et al PRECLINICAL LENTIVIRAL RAG1 GENE THERAPY Supplemental Figure 3 7 PIKE-OVERZET et al PRECLINICAL LENTIVIRAL RAG1 GENE THERAPY Supplemental References 1. Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, et al. A thirdgeneration lentivirus vector with a conditional packaging system. J Virol. 1998 Nov;72(11):8463-71. 2. Schambach A, Bohne J, Baum C, Hermann FG, Egerer L, von Laer D, et al. Woodchuck hepatitis virus post-transcriptional regulatory element deleted from X protein and promoter sequences enhances retroviral vector titer and expression. Gene Ther. 2006 Apr;13(7):641-5. 3. Charrier S, Stockholm D, Seye K, Opolon P, Taveau M, Gross DA, et al. A lentiviral vector encoding the human Wiskott-Aldrich syndrome protein corrects immune and cytoskeletal defects in WASP knockout mice. Gene Ther. 2005 Apr;12(7):597-606. 4. Sirven A, Ravet E, Charneau P, Zennou V, Coulombel L, Guetard D, et al. Enhanced transgene expression in cord blood CD34(+)-derived hematopoietic cells, including developing T cells and NOD/SCID mouse repopulating cells, following transduction with modified trip lentiviral vectors. Mol Ther. 2001 Apr;3(4):438-48. 8