Supplementary Information Linoleic acid metabolite drives severe
advertisement

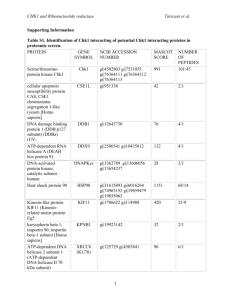
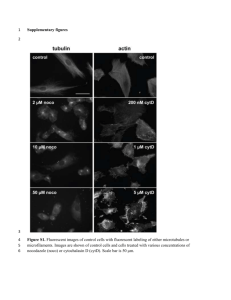
Supplementary Information Linoleic acid metabolite drives severe asthma by inducing mitochondrial dysfunction and airway epithelial injury Ulaganathan Mabalirajan1*, Rakhshinda Rehman1, Tanveer Ahmad1, Sarvesh Kumar2, Sucitha Singh1, Geeta D. Leishangthem3, Jyotirmoi Aich1, Manish Kumar1, Kritika Khanna1, Vijay P. Singh1, Amit K. Dinda3, Shyam Biswal2, Anurag Agrawal1, and Balaram Ghosh1 1Molecular Immunogenetics Laboratory and Centre of Excellence for Translational Research in Asthma and Lung Disease, CSIR-Institute of Genomics and Integrative Biology, India. 2Department of Environmental Health Sciences, Bloomberg School of Public Health, Johns Hopkins University, USA. 3Division of Renal Pathology, Department of Pathology, All India Institute of Medical Sciences, India. *Corresponding Author: Ulaganathan Mabalirajan, Molecular Immunogenetics Laboratory, CSIR-Institute of Genomics and Integrative Biology, Mall Road, Delhi-110 007. Phone: 91-11-27662580. Fax: 91-1127667471. E-mail address: um.rajan@igib.res.in Supplementary Figure S1. Murine models used in this study. a) Different concentrations of 13-S-hydroxyoctadecadienoic acid (13-S-HODE) or vehicle (0-0.6–mg dose per mouse in BALB/c mice or 0.6-mg dose per mouse in C57BL/6 mice) were instilled intranasally on days 1 to 3 in isoflurane-anesthetized mice. Respiratory mechanics was measured, and euthanasia was performed 6 hours after the last dose. Intraperitoneal administration of 0.05 mg/kg calpeptin (CALP) or 2.5 mg/kg capsazepine (CPZ) or vehicle or intranasal administration of Transient Receptor Potential Cation Channel, Vanilloid-type 1 (TRPV1) siRNA or scrambled siRNA was performed in mice in which 13-S-HODE had been administered. b) BALB/c mice were sensitized on days 0, 7, and 14, followed by seven challenges with OVA, and AHR was measured and euthanasia was performed 24 hours after the last challenge. 13-S-HODE antibody or isotype control was administered intraperitoneally on days 25 to 27 in BALB/c mice. TRPV1 siRNA, or scrambled siRNA was administered intranasally on days 25 and 27 in BALB/c mice. CPZ (2.5 mg/kg) or VEH twice a day was administered intraperitoneally on days 21 to 27 in BALB/c mice. AHR, airway hyperresponsiveness; VEH, vehicle. Supplementary Figure S2. Effect of ethanol on normal airways. To determine the possible toxic effects of vehicle in which 13-S-HODE was dissolved (50 % ethanol), histopathology of alveolar regions and baseline lung compliance of vehicle administered BALB/c mice were estimated and compared with normal control mice. a) Photomicrographs of hematoxylin and eosin stained alveolar regions of normal mice and mice which were administered 50 % ethanol intranasally once a day for 3 consecutive days followed by sacrifice 6 hours after the last administration (n = 5-6 mice per group). All images are at 20X magnification. Invasive measurements to determine baseline lung compliance (b) and lung elastance in different groups of BALB/c mice (c). Control, naïve mice which were not given either vehicle or 13-S-HODE. Supplementary Figure S3. 13-S-HODE–induced mitochondrial dysfunction and epithelial injury seem to be mediated by TRPV1. a) Inflammation scoring was performed in lung sections stained with hematoxylin and eosin from mice in which 13-S-HODE was administered and that were treated with CPZ (a pharmacological antagonist of TRPV1) n = 6 mice per group). b) Graph shows data from ELISA of 8-isoprostane, a marker of oxidative stress. c) Graph shows data from photomicrographs of TUNEL-stained lung sections that were assessed for the percentage of TUNEL-positive bronchial epithelium. d) Inflammation scoring was performed in lung sections stained with hematoxylin and eosin from mice in which 13-S-HODE was administered and that were treated with either TRPV1 siRNA or scrambled siRNA (n = 4-6 mice per group). e) Graph shows data from ELISA of 8-isoprostane, a marker of oxidative stress in lung cytosol. f) Differential cell count was performed in BAL fluid and expressed as the percentage of each cell type. Results are shown as mean ± s.e.m., significance is determined with unpaired Student t test or Mann-Whitney test (*P ˂ 0.0001; ***P ˂ 0.05). Macro, macrophage; Mono, mononuclear agranulocytes such as monocytes and lymphocytes; and Neutro, neutrophil. Supplementary Figure S4. Pharmacological antagonism of TRPV1 attenuates asthma features, restores mitochondrial dysfunction, and reduces epithelial injury in allergic mice. a) Graph of inflammation score derived from photomicrographs of bronchovascular regions stained with hematoxylin and eosin from mice that were sensitized and challenged with alum or OVA for 7 consecutive days and administered either CPZ or vehicle (n = 6 mice per group). b) Differential cell count was performed in BAL fluid and expressed as the percentage of each cell type. Macro, macrophage; Mono, mononuclear agranulocytes such as monocytes and lymphocytes; Eosino, eosinophil; and Neutro, neutrophil. c) Activity of mitochondrial complex I (n = 4-6 mice per group). (d, e) Graphs of data from ELISA and fluorescence-based activity assays showed cytochrome c and caspase 3 activity in lung cytosol. Results shown as mean ± s.e.m., significance determined with unpaired Student t test or Mann-Whitney test (***P ˂ 0.05). SHAM/PBS/VEH, PBS–sensitized, and PBS-challenged mice treated with vehicle; OVA/OVA/VEH, OVA–sensitized, and OVA-challenged mice treated with vehicle; and OVA/OVA/CPZ, OVA–sensitized, and OVA-challenged mice treated with capsazepine. Supplementary Table 1. Symbol Mitochondrial functions Mitochondrial dynamics Mfn1 Mfn2 Msto1 Dnm1l Fis1 Opa1 Fold change vs. HODE Fold change vs. OVA Gene name 1.5227 -1.3044 2.4061 1.8747 1.4406 1.5547 1.1514 -1.3074 -1.1947 -1.0994 1.1837 1.2002 Uncoupling Ucp2 Ucp3 3.6723 -1.2086 2.3349 Uncoupling protein 2 (mitochondrial, proton carrier) -1.0257 Uncoupling protein 3 (mitochondrial, proton carrier) Chaperones and antioxidants Hsp90aa1 Hspd1 Sod1 Sod2 -1.651 -1.5298 1.4109 1.1701 -1.0842 1.217 -2.214 1.3692 Heat shock Heat shock Superoxide Superoxide Solute carriers Slc25a1 Slc25a10 Slc25a20 Slc25a21 Slc25a22 Slc25a15 Slc25a2 Slc25a12 Slc25a13 Slc25a3 Slc25a23 Slc25a24 Slc25a19 Slc25a25 Slc25a4 Slc25a5 Slc25a14 Slc25a16 Slc25a17 Slc25a27 Slc25a30 Slc25a31 Slc25a37 1.4607 1.3074 2.4566 -1.7451 1.2114 2.0515 1.3819 2.3403 2.4228 1.8747 1.1865 3.2641 -1.4273 1.2198 -1.1355 1.4709 1.0619 2.3729 1.9274 2.3403 1.8747 2.3894 2.0373 1.488 -1.7132 1.1837 -3.1529 1.5404 1.3503 -1.4406 1.9498 1.5948 1.2953 -1.9141 1.8965 -1.0046 -1.6896 -1.2541 1.5298 -1.2114 1.0163 1.0817 -1.2983 1.0521 -1.6434 -1.2283 Solute Solute Solute Solute Solute Solute Solute Solute Solute Solute Solute Solute Solute Solute Solute Solute Solute Solute Solute Solute Solute Solute Solute Mitochondrial import machinery Fxc1 Timm10 Timm17a Timm17b Timm22 Timm23 Timm44 Timm50 Timm8a1 Timm8b Timm9 Grpel1 Dnajc19 Tomm20 Tomm22 Tomm34 Tomm40 Tomm40l Tomm70a Mtx2 Sh3glb1 Cox18 Aip Immp1l Immp2l Mipep 1.0619 1.0994 1.3441 1.0767 1.7613 -1.1045 1.6208 1.9141 -1.5619 1.0546 -1.234 2.0515 1.7492 1.7736 -1.3787 -1.3044 1.5122 1.146 1.1147 -1.2953 -1.1674 1.7251 1.154 -1.0668 -1.674 -1.1755 1.6283 -1.2894 -1.2894 1.0163 -1.1381 1.0817 -1.1147 1.8575 1.0817 -1.4208 -1.4208 2.4852 1.1199 -1.3915 -1.8234 -2.2294 -1.0918 1.7695 1.1199 1.0668 -1.2198 2.434 1.2086 -1.2894 -1.0046 -1.3629 Fractured callus expressed transcript 1 Translocase of inner mitochondrial membrane 10 homolog (yeast) Translocase of inner mitochondrial membrane 17a Translocase of inner mitochondrial membrane 17b Translocase of inner mitochondrial membrane 22 homolog (yeast) Translocase of inner mitochondrial membrane 23 homolog (yeast) Translocase of inner mitochondrial membrane 44 Translocase of inner mitochondrial membrane 50 homolog (yeast) Translocase of inner mitochondrial membrane 8 homolog a1 (yeast) Translocase of inner mitochondrial membrane 8 homolog b (yeast) Translocase of inner mitochondrial membrane 9 homolog (yeast) GrpE-like 1, mitochondrial DnaJ (Hsp40) homolog, subfamily C, member 19 Translocase of outer mitochondrial membrane 20 homolog (yeast) Translocase of outer mitochondrial membrane 22 homolog (yeast) Translocase of outer mitochondrial membrane 34 Translocase of outer mitochondrial membrane 40 homolog (yeast) Translocase of outer mitochondrial membrane 40 homolog-like (yeast) Translocase of outer mitochondrial membrane 70 homolog A (yeast) Metaxin 2 SH3-domain GRB2-like B1 (endophilin) COX18 cytochrome c oxidase assembly homolog (S. cerevisiae) Aryl-hydrocarbon receptor-interacting protein IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) Mitochondrial intermediate peptidase Mitochondrial trafficking Rhot1 Rhot2 Lipid and heme metabolism Stard3 Tspo Cpt2 Cpt1b Taz Cox10 2.4737 1.3256 1.1865 1.2198 1.5984 7.1437 1.9954 -1.8067 RNA stabiity Lrpprc Uxt 1.0767 1.3074 Cell survival Cdkn2a Sfn Pmaip1 Trp53 Aifm2 Bid Bnip3 Bak1 Bbc3 Bcl2 Bcl2l1 -1.0892 -1.341 2.0946 -1.398 1.0329 1.154 4.3974 1.1947 1.7132 1.2541 3.7494 Nefl 2.5082 Mitofusin 1 Mitofusin 2 Misato homolog 1 (Drosophila) Dynamin 1-like Fission 1 (mitochondrial outer membrane) homolog (yeast) Optic atrophy 1 homolog (human) protein 90, alpha (cytosolic), class A member 1 protein 1 (chaperonin) dismutase 1, soluble dismutase 2, mitochondrial carrier carrier carrier carrier carrier carrier carrier carrier carrier carrier carrier carrier carrier carrier carrier carrier carrier carrier carrier carrier carrier carrier carrier family family family family family family family family family family family family family family family family family family family family family family family 25 (mitochondrial 25 (mitochondrial 25 (mitochondrial 25 (mitochondrial 25 (mitochondrial 25 (mitochondrial 25 (mitochondrial 25 (mitochondrial 25 (mitochondrial 25 (mitochondrial 25 (mitochondrial 25 (mitochondrial 25 (mitochondrial 25 (mitochondrial 25 (mitochondrial 25 (mitochondrial 25 (mitochondrial 25 (mitochondrial 25 (mitochondrial 25, member 27 25, member 30 25 (mitochondrial 25, member 37 carrier, citrate transporter), member 1 carrier, dicarboxylate transporter), member 10 carnitine/acylcarnitine translocase), member 20 oxodicarboxylate carrier), member 21 carrier, glutamate), member 22 carrier ornithine transporter), member 15 carrier, ornithine transporter) member 2 carrier, Aralar), member 12 carrier, adenine nucleotide translocator), member 13 carrier, phosphate carrier), member 3 carrier; phosphate carrier), member 23 carrier, phosphate carrier), member 24 thiamine pyrophosphate carrier), member 19 carrier, phosphate carrier), member 25 carrier, adenine nucleotide translocator), member 4 carrier, adenine nucleotide translocator), member 5 carrier, brain), member 14 carrier, Graves disease autoantigen), member 16 carrier, peroxisomal membrane protein), member 17 carrier; adenine nucleotide translocator), member 31 1.5087 Ras homolog gene family, member T1 -1.0767 Ras homolog gene family, member T2 1.3044 1.4473 1.1594 2.835 -1.4406 1.0023 START domain containing 3 Translocator protein Carnitine palmitoyltransferase 2 Carnitine palmitoyltransferase 1b, muscle Tafazzin COX10 homolog, cytochrome c oxidase assembly protein, heme A: farnesyltransferase (yeast) -1.0994 Leucine-rich PPR-motif containing 1.674 Ubiquitously expressed transcript 1.8965 -3.3096 1.1199 1.0595 -1.6096 1.923 -1.5547 1.3692 -1.0693 2.2868 2.2243 Cyclin-dependent kinase inhibitor 2A Stratifin Phorbol-12-myristate-13-acetate-induced protein 1 Transformation related protein 53 Apoptosis-inducing factor, mitochondrion-associated 2 BH3 interacting domain death agonist BCL2/adenovirus E1B interacting protein 3 BCL2-antagonist/killer 1 BCL2 binding component 3 B-cell leukemia/lymphoma 2 Bcl2-like 1 5.7491 Neurofilament, light polypeptide Suppl. Table 1. Quantitative real-time PCR array profiling of key genes of mitochondrial functions shows the differentially expressed genes in murine lung samples in which 13-S-HODE was administered and or allergic mice. OVA-induced asthmatic mice had common upregulated genes such as Ucp2, GrPel1, Cpt1b, Bcl2l, and Nefl compared with findings in 13-S-HODE–induced asthmatic mice. More than or less than twofold increase or decrease indicates differentially expressed genes compared to either SHAM control mice or vehicle administered mice. Supplementary Table 2. Symbol Fold change Description Complex I Core Core Core Core Core Core Core Regulation of complex I activity Supernumerary Supernumerary Supernumerary Supernumerary Supernumerary Supernumerary Supernumerary Accessory subunit Accessory subunit Accessory subunit Accessory subunit Accessory subunit Accessory subunit Accessory subunit Accessory subunit Accessory subunit Accessory subunit Accessory subunit Accessory subunit Accessory subunit Accessory subunit Accessory subunit Accessory subunit Accessory subunit Accessory subunit Ndufs7 Ndufs8 Ndufv1 Ndufv2 Ndufs1 Ndufs2 Ndufs3 Ndufs4 Ndufa1 Ndufa10 Ndufa11 Ndufa2 Ndufab1 Ndufb9 Ndufs6 Ndufa3 Ndufa4 Ndufa5 Ndufa6 Ndufa7 Ndufa8 Ndufb10 Ndufb2 Ndufb3 Ndufb4 Ndufb5 Ndufb6 Ndufb7 Ndufb8 Ndufc1 Ndufc2 Ndufs5 Ndufv3 -1.3379 -1.4743 -1.2924 -1.4641 1.0497 -1.2397 1.2311 1.0353 1.5476 1.8277 1.5911 -1.8404 -1.5369 1.4044 -1.257 -1.6818 -1.2658 -1.1408 1.2311 -1.2058 1.007 1.1096 -1.0718 -1.1019 -1.1892 -1.2834 -1.1975 -1.2483 -1.0867 -1.1251 1.0425 -1.1251 -1.257 NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH NADH dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase dehydrogenase (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) (ubiquinone) Fe-S protein 7 Fe-S protein 8 flavoprotein 1 flavoprotein 2 Fe-S protein 1 Fe-S protein 2 Fe-S protein 3 Fe-S protein 4 1 alpha subcomplex, 1 1 alpha subcomplex 10 1 alpha subcomplex 11 1 alpha subcomplex, 2 1, alpha/beta subcomplex, 1 1 beta subcomplex, 9 Fe-S protein 6 1 alpha subcomplex, 3 1 alpha subcomplex, 4 1 alpha subcomplex, 5 1 alpha subcomplex, 6 (B14) 1 alpha subcomplex, 7 (B14.5a) 1 alpha subcomplex, 8 1 beta subcomplex, 10 1 beta subcomplex, 2 1 beta subcomplex 3 1 beta subcomplex 4 1 beta subcomplex, 5 1 beta subcomplex, 6 1 beta subcomplex, 7 1 beta subcomplex 8 1, subcomplex unknown, 1 1, subcomplex unknown, 2 Fe-S protein 5 flavoprotein 3 Sdha Sdhb Sdhc Sdhd 1.5052 1.6358 -1.3379 1.2658 Succinate Succinate Succinate Succinate Uqcrc1 Uqcrc2 Cyc1 Uqcrfs1 Uqcr11 Uqcrh Uqcrq Bcs1l 1.0644 -1.1647 -1.3566 1.2658 -1.5583 -1.1728 -1.2746 1.0644 Ubiquinol-cytochrome Ubiquinol cytochrome Cytochrome c-1 Ubiquinol-cytochrome Ubiquinol-cytochrome Ubiquinol-cytochrome Ubiquinol-cytochrome BCS1-like (yeast) Cox4i1 Cox4i2 Cox5a Cox5b Cox6a1 Cox6a2 Cox6b1 Cox6b2 Cox6c Cox7a2 Cox7a2l Cox7b Cox8a Cox8c Oxa1l Cox11 1.1019 1.815 -2.395 -1.0867 1.3195 1.1019 1.2311 -1.3755 -1.0943 -1.1892 -1.1487 -1.5692 -1.0644 1.1892 1.6245 -1.434 Cytochrome c oxidase subunit IV isoform 1 Cytochrome c oxidase subunit IV isoform 2 Cytochrome c oxidase, subunit Va Cytochrome c oxidase, subunit Vb Cytochrome c oxidase, subunit VI a, polypeptide 1 Cytochrome c oxidase, subunit VI a, polypeptide 2 Cytochrome c oxidase, subunit VIb polypeptide 1 Cytochrome c oxidase subunit VIb polypeptide 2 Cytochrome c oxidase, subunit VIc Cytochrome c oxidase, subunit VIIa 2 Cytochrome c oxidase subunit VIIa polypeptide 2-like Cytochrome c oxidase subunit VIIb Cytochrome c oxidase, subunit VIIIa Cytochrome c oxidase, subunit VIIIc Oxidase assembly 1-like COX11 homolog, cytochrome c oxidase assembly protein (yeast) 1.1892 1.0497 1.1892 -2.0849 1.2397 1.4948 -2.5847 -1.4641 1.3379 1.4241 1.8404 1.815 -1.7171 -1.2142 1.0353 1.5476 1.0425 -1.5583 1.1892 1.1892 ATPase, H+/K+ transporting, nongastric, alpha polypeptide ATPase, H+/K+ exchanging, gastric, alpha polypeptide ATPase, H+/K+ exchanging, beta polypeptide ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit ATP synthase, H+ transporting, mitochondrial F0 complex, subunit B1 ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c1 (subunit 9) ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 (subunit 9) ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F2 ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit ATPase, H+ transporting, lysosomal V0 subunit A2 ATPase, H+ transporting, lysosomal V0 subunit D2 ATPase, H+ transporting, lysosomal V1 subunit C2 ATPase, H+ transporting, lysosomal V1 subunit E2 ATPase, H+ transporting, lysosomal V1 subunit G3 Complex II Catalytic site Catalytic site Membrane-anchoring subunit Membrane-anchoring subunit dehydrogenase dehydrogenase dehydrogenase dehydrogenase complex, complex, complex, complex, subunit subunit subunit subunit A, B, C, D, flavoprotein (Fp) iron sulfur (Ip) integral membrane protein integral membrane protein Complex III Core subunit Core subunit Respiratory Respiratory Low molecular weight Low molecular weight Low molecular weight assembly of complex subunit subunit subunit III c reductase core protein 1 c reductase core protein 2 c c c c reductase, Rieske iron-sulfur polypeptide 1 reductase, complex III subunit XI reductase hinge protein reductase, complex III subunit VII Complex IV regulation and assembly regulation and assembly regulation and assembly regulation and assembly regulation and assembly regulation and assembly regulation and assembly Connects the two COX monomers regulation and assembly regulation and assembly regulation and assembly regulation and assembly regulation and assembly regulation and assembly activity and assembly of cytochrome oxidase insertion of the copper B into subunit I Complex V catalytic subunit of the ouabain-sensitive H+/K+ -ATPase Atp12a catalytic alpha subunit of the gastric H+, K+-ATPase Atp4a catalytic beta subunit of the gastric H+, K+-ATPase Atp4b alpha subunit of the catalytic core Atp5a1 beta subunit of the catalytic core Atp5b gamma subunit of the catalytic core Atp5c1 delta subunit of the catalytic core Atp5d b subunit of the proton channel Atp5f1 subunit c of the proton channel Atp5g1 subunit c of the proton channel Atp5g2 subunit c of the proton channel Atp5g3 d subunit of the proton channel Atp5h F6 subunit of proton channel Atp5j f subunit of the proton channel Atp5j2 part of the connector linking catalytic core and proton channel Atp5o component of the V(0) domain vacuolar ATPase Atp6v0a2 Subunit of the integral membrane V0 complex of vacuolar ATPase Atp6v0d2 Subunit of the peripheral V1 complex of vacuolar ATPase Atp6v1c2 Subunit of the peripheral V1 complex of vacuolar ATPase Atp6v1e2 Catalytic subunit of peripheral V1 vacuolar ATPase Atp6v1g3 other nuclear coded OXPHOS genes Lhpp Ppa1 Ppa2 1.0943 Phospholysine phosphohistidine inorganic pyrophosphate phosphatase 1.0792 Pyrophosphatase (inorganic) 1 -1.5583 Pyrophosphatase (inorganic) 2 Supplementary Table 2. Quantitative real-time PCR array profiling of key genes of the mitochondrial electron transport chain shows the differentially expressed genes in lungs in which 13-S-HODE was administered. Downregulated genes are in blue, and unaltered genes are in black; more than or less than twofold increase or decrease indicates differentially expressed genes compared to either SHAM control mice or vehicle administered mice. Legends for supplementary videos: 1. Supplementary Video 1 (MOV 162 K) Breathing pattern of mouse in which VEH (ethanol) was administered. The breathing pattern was captured 10 to 15 minutes after administration of 30 µl of ethanol in isofluraneanesthetized BALB/c mice. 2. Supplementary Video 2 (MOV 231 K) Breathing pattern of mouse in which 13-S-HODE was administered. The breathing pattern was captured 10 to 15 minutes after administration of 30 µl of 13-S-HODE (0.6 mg/d per mouse for 3 d) in isoflurane-anesthetized BALB/c mice. The moving mouse with the yellow label was administered VEH. There was visible difficulty in breathing characterized by forced abdominal movement with complete focus on breathing in 13-S-HODE–treated mice compared with no focus on breathing in ethanol-treated mice. 3. Supplementary Video 3 (MOV 110 K) Breathing pattern of mouse in which 13-S-HODE was administered. The breathing pattern was captured 10 to 15 minutes after administration of 30 µl of 13-S-HODE (0.6 mg/d per mouse for 3 d) in isoflurane-anesthetized BALB/c mice that were pretreated with 2.5 mg/kg CPZ. 4. Supplementary Video 4 (MOV 327 K) Breathing pattern of mouse in which 13-S-HODE was administered. The breathing pattern was captured 10 to 15 minutes after administration of 30 µl of 13-S-HODE (0.6 mg/d per mouse for 3 d) in isoflurane-anesthetized BALB/c mice that were pretreated with 50 µg of TRPV1 siRNA per day per mouse.