jbi12487-sup-0002-AppendixS2
advertisement

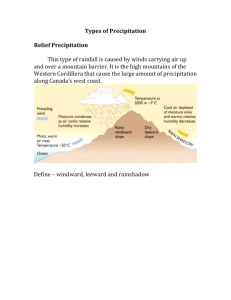
SUPPORTING INFORMATION – Journal of Biogeography The interplay of spatial and climatic landscapes in the genetic distribution of a South American parrot Juan F. Masello, Valeria Montano, Petra Quillfeldt, Soňa Nuhlíčková, Martin Wikelski and Yoshan Moodley Appendix S2 Supplementary figures (Figs S1–S10) Figure S1 Plot of the matrix of correlations for latitude, longitude and the 21 climatic variables from the studied burrowing parrot (Cyanoliseus patagonus) localities. Variable abbreviations: Mean monthly minimum Temperature (MMminT), Mean monthly maximum Temperature (MMmaxT), Annual Mean Temperature (MAT), Mean Monthly Temperature Range (MMTR), Isothermality (Iso), Temperature Seasonality (TS), Maximum Temperature of Warmest Month (MaxTWaM), Minimum Temperature of Coldest Month (MinTCoM), Temperature Annual Range (TAR), Mean Temperature of Wettest Quarter (MTWetQ), Mean Temperature of Driest Quarter (MTDriQ), Mean Temperature of Warmest Quarter (MTWaQ), Mean Temperature of Coldest Quarter (MTCoQ), Annual Precipitation (AP), Precipitation of Wettest Month (PWetM), Precipitation of Driest Month (PDriQ), Precipitation Seasonality (PS), Precipitation of Wettest Quarter (PWetQ), Precipitation of Driest Quarter (PDriQ), Precipitation of Warmest Quarter (PWaQ), Precipitation of Coldest Quarter (PCoQ). Temperature Seasonality (TS): the standard deviation of the weekly mean temperatures expressed as a percentage of the mean of those temperatures (i.e. the annual mean). Isothermality (Iso): the mean diurnal range divided by the Annual Temperature Range. Precipitation Seasonality (PS): the standard deviation of the weekly precipitation estimates expressed as a percentage of the mean of those estimates (i.e. the annual mean). See also Supplementary Table S3. sPCA Eigenvalues Figure S2 Connection network (a), bar plots showing each component ordered by eigenvalue (b), and plots of the three components with highest positive eigenvalues (ce) after sPCA based on temperature (TPC-1 and TPC-2; P = 0.025) at all sampled burrowing parrot localities. Numbers represent the localities and are given as in Table 2. For a plot showing the eigenvalue of each component (y-axis) and the amount of genetic variance it described (x-axis) see Fig. S7. Colonies are colour coded as in Fig. 2. For a detailed interpretation of the black and white squares and their size see the legend of Fig. 2. TPC: Temperature Principal Component. Figure S3 Connection network (a), bar plots showing each component ordered by eigenvalue (b), and plots of the three components with highest positive eigenvalues (ce) after sPCA based on precipitation (PPC-2 and PPC-3; P = 0.023) at all sampled burrowing parrot localities. Numbers represent the localities and are given as in Table 2. For a plot showing the eigenvalue of each component (y-axis) and the amount of genetic variance it described (x-axis) see Fig. S8. Colonies are colour coded as in Fig. 2. For a detailed interpretation of the black and white squares and their size see the legend of Fig. 2. PPC: Precipitation Principal Component. Figure S4 Connection network (a), bar plots showing each component ordered by eigenvalue (b), and plots of the three components with highest positive eigenvalues (ce) after sPCA based on precipitation (PPC-3 and PPC-4; P = 0.033) at all sampled burrowing parrot localities. Numbers represent the localities and are given as in Table 2. For a plot showing the eigenvalue of each component (y-axis) and the amount of genetic variance it described (x-axis) see Fig. S9. Colonies are colour coded as in Fig. 2. For a detailed interpretation of the black and white squares and their size see the legend of Fig. 2. PPC: Precipitation Principal Component. Figure S5 Connection network (a), bar plots showing each component ordered by eigenvalue (b), and plots of the three components with highest positive eigenvalues (ce) after sPCA based on precipitation (PPC-3 and PPC-5; P = 0.007) at all sampled burrowing parrot localities. Numbers represent the localities and are given as in Table 2. For a plot showing the eigenvalue of each component (y-axis) and the amount of genetic variance it described (x-axis) see Fig. S10. Colonies are colour coded as in Fig. 2. For a detailed interpretation of the black and white squares and their size see the legend of Fig. 2. PPC: Precipitation Principal Component. Figure S6 The eigenvalue of each component (y-axis) and the amount of genetic variance it described (x‐axis) for geography (latitude plus longitude). Figure S7 The eigenvalue of each component (y-axis) and the amount of genetic variance it described (x‐axis) for TPC‐1 and TPC‐2. TPC: Temperature Principal Component. Figure S8 The eigenvalue of each component (y-axis) and the amount of genetic variance it described (x‐axis) for PPC‐2 and PPC‐3. PPC: Precipitation Principal Component. Figure S9 The eigenvalue of each component (y-axis) and the amount of genetic variance it described (x‐axis) for PPC‐3 and PPC‐4. PPC: Precipitation Principal Component. Figure S10 The eigenvalue of each component (y-axis) and the amount of genetic variance it described (x‐axis) for PPC‐3 and PPC‐5. PPC: Precipitation Principal Component.