Real-time quantitative reverse transcription PCR (RT-qPCR)
advertisement

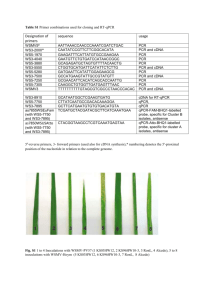
Materials and methods Human samples Skin biopsies from 52 consenting CMT1A patients enrolled in the ascorbic acid italian trial were obtained from the medial aspect of proximal phalanx of the index (Milan and Genoa centres) or from the tip of the little finger (Naples) and studied. The procedure was performed using a 3 mm biopsy punch under sterile conditions and local anaesthesia with 1% lidocaine. Seven pairs of skin biopsies, which were known to have encountered problems of cryopreservation, did not reach the minimum standard quality of total RNA and were then discarded. Thus, the 45 remaining patients were analysed in this study. To evaluate splicing variants tissue distribution, we used Human I MTC Panels (Clontech, USA) containing cDNAs derived from heart, brain, placenta, lung, liver, skeletal muscle, kidney and pancreas human tissues. Primary human fibroblast cultures and sorted human lymphocytes were also used. Absolute quantification of PMP22 mRNA In order to verify whether skin biopsy is a suitable tissue to measure PMP22 mRNA levels, we cloned full length cDNA of the human PMP22 splicing variants in pDrive Cloning Vector through Qiagen PCR cloning Plus Kit (Qiagen, Italy). The sequences of each plasmid were verified through sequencing (TibMolBiol, Italy) and are available upon request. Plasmids containing cDNA of each PMP22 splicing variants were used to perform a titration curve assay. Pure plasmid preparations (Qiaprep spin miniprep kit, Qiagen, Italy) were gradually diluted and utilised as standard positive controls in Real-Time quantitative PCR (RT-qPCR) reactions to calculate the absolute copy number of PMP22 mRNA in skin biopsies and to test PCR reaction efficiency of our primers (see next paragraph). Real-time quantitative reverse transcription PCR (RT-qPCR) Human skin and sural nerve biopsies were homogenised in Qiazol reagent through a high speed shaking Tissue Lyser II system using the 5 mm stainless steel beads (Qiagen s.r.l. - Italy). Paired samples (T0 and T24 skin biopsies from the same patient and the age and gender- matched normal subject), were processed at the same time to minimise experimental errors and handling. RNA extraction from tissues was performed with miRNeasy mini kit (Qiagen-Italy) according to manufacturer’s protocols. Total RNA was resuspended in RNase free water and checked for quality and quantity by Nanodrop technology (NanoDrop products, Wilmington, DE, USA). The quality of primers used in this study was checked for intron spanning and matching common PCR standard parameters by BLASTN algorithm. Specificity and efficiency of the amplification product for each primer pair was verified by the presence of a single peak in RT-qPCR melting curve and by the achievement to the cut-off value through a titration curve. We have utilised multiple reference genes to normalise PMP22 mRNA according to MIQE standard guidelines for RT-qPCR (Bustin et al. , 2010). The most stable three reference genes were chosen from a pool of seven candidate genes deriving from independent pathways and tested for intergroup stability in our skin and nerve samples using geNorm algorithm (Vandesompele et al. , 2002). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), succinate dehydrogenase complex, subunit A, flavoprotein (SDHA), and TATA box binding protein (TBP) reached a coefficient of stability (M value) lower than the cut-off value, and they were therefore selected for the RT-qPCR experiments. Ct data (cycle thresold) were calculated with ΔCt method using as internal calibrator the sample with lowest Ct and resulting values were analysed by geNorm algorithm for normalization. Finally, the variability of each sample has been calculated to verify the reproducibility of our measurements. All PCR reactions were carried out in duplicate and averaged. Statistical analysis Reproducibility and repeatability of splicing variants measurements were inspected by the BlandAltman plot and analyzed with the Friedman’s repeated measures ANOVA on ranks. References Bustin SA, Beaulieu JF, Huggett J, Jaggi R, Kibenge FS, Olsvik PA, et al. MIQE precis: Practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol Biol 2010; 11: 74. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002; 3: RESEARCH0034.