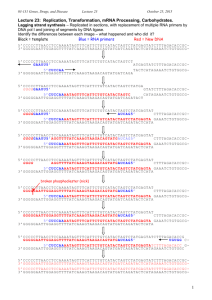
Supplementary material Q/R site Bbv1 restriction analysis The e
advertisement

Supplementary material Q/R site Bbv1 restriction analysis The extents of RNA editing at the Q/R site in GluA2 mRNA and pre-mRNA were expressed as the percentage of edited transcripts in the total transcripts. This was calculated by quantitative analyses of the restriction digests of PCR products with optical density analysis using Gel Pro Analyzer software. Digestion of the PCR products with BbvI yielded two bands at 245 and 76 bp when they originated from edited GluA2 mRNA, whereas three bands at 106, 139 and 76 bp when they originated from unedited GluA2 mRNA. The 76 bp band originated from both edited and unedited GluA2 mRNA, while the 245 bp band originated only from edited mRNA, hence the extent of editing was calculated as the molarity of the former to the latter for each sample. With regard to the pre-mRNA, the digestion of the PCR products with BbvI did not cut the edited GluA2 pre-mRNA leaving a PCR product of 245 bp, while it yielded two bands at 139 and 105 bp when they originated from unedited GluA2. Here the extent of editing was calculated as the molarity of the 245 bp band (edited transcripts) against 245 bp + 139 bp (total transcripts) molarities. Relative quantification of ADAR2 mRNA isoforms Analysis of rat ADAR2 alternative splicing products was performed after RT-PCR amplification of total RNA from each sample. Using a primer pair located in exon 1 and in exon 4 relative to the rat sequence (NM_001111056), both the normal and the alternative spliced (+47nt) transcripts were detected 1. In addition, using primers located on exon 7 and exon 8, we analyzed the splicing pattern involving the deaminase domain which leads to the formation of the +30/-30 isoforms. The Agilent 2100 bioanalyzer was used to analyze alternatively spliced ADAR2 isoforms2. The bioanalyzer uses lab-on-a-chip technology to perform gel electrophoresis. Samples were separated electrophoretically in a polymer solution, similar to capillary electrophoresis. This instrument detects laserinduced fluorescence using an intercalating dye, which is added to the polymer. The bioanalyzer software automatically calculates the size and concentration of each separate band. The PCR products were analyzed using DNA 1000 Lab kit chips (Agilent Technologies, Waldbronn, Germany). All the chips were prepared according to the manufacturer’s instructions. Western blot analysis Cells harvested from primary cortical cultures were solubilized with modified RIPA (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% IGEPAL CA630, 0.25% NaDOC, 0.1% SDS, 1% NP-40 and Roche protease inhibitor tablets) and then sonicated. A portion of the lysate was used for the Bicinchoninic Acid (BCA) protein concentration assay (SigmaAldrich). Equal amounts of protein were applied to precast SDS polyacrylamide gels (4– 12% NuPAGE Bis-Tris gels; Invitrogen) and the proteins were electrophoretically transferred to a Hybond-P PVDF membrane (GE Healthcare) Membrane (GE Healthcare, Waukesha, WI, USA) for 2 h. The membranes were blocked for 60 min with 3% nonfat dry milk in TBS-T (Tris-buffered saline with 0.1% Tween-20, Sigma-Aldrich) and then incubated overnight at 4°C in the blocking solution with the mouse monoclonal anti-aIISpectrin (1:2000; Enzo Life Sciences. Cod: MBL-FG6090). For detection, after three washes in TBS-T, the membranes were incubated for 1 h at room temperature. The membranes were subsequently incubated for one hour at RT, with AP-conjugated secondary antibodies (Santa Cruz Biotechnology), and then incubated with CDPstar blotting detection reagents (Roche Applied Science). Supplemental Results Fig. S1: Analysis of Q/R editing site on GluA2 pre-mRNA and mRNA with two different techniques. Direct sequencing was performed to determine the percentage of Q/R site-edited GluA2 both on pre-mRNA (A) and on mRNA (B). The level of editing was around 88% by analyzing GluA2 pre-mRNA and mostly fully edited by analyzing the GluA2 mRNA, thus indicating that the unedited transcript might be not spliced. However 24h of glutamate treatment did not modify the editing level either at the pre-mRNA or mRNA levels. BbvI digestion was carried out on the PCR product obtained from pre-mRNA (C) and mRNA (D) and restriction digested products were loaded on an agarose gel. Control samples were loaded in lanes 1-2-3, 24hr glutamate treated samples in lanes 4-5-6, undigested PCR product was loaded in lane 7, and the marker in lane 8. Quantifications of Q/R-edited GluA2 in GluA2 pre-mRNA (E) and mRNA (F) were expressed as the ratio (%) of edited transcripts in total transcripts, as reported in the supplemental material. BbvI digestion led to results similar to those from the sequencing analysis. GluA2 Q/R site was not fully edited at the pre-mRNA level (nearly 85%), while it was almost 100% at the mRNA level. 24h glutamate treatment did not alter the GluA2 Q/R editing level. (G) Scheme for GluA2 pre-mRNA, showing size of PCR product (244bp) and the sizes of BbvI fragments for unedited GluA2 (139bp and 105bp). (H) Scheme for mRNA, showing size of PCR product (321bp) and the sizes of BbvI fragments for edited GluA2 (245bp and 76bp) and unedited GluA2 (139bp, 105bp and 76bp). Fig. S2: Effect of chronic glutamate treatment on ADAR2 splicing pattern. (A) Analysis of ADAR2 alternative splicing at the 47nt cassette and (C) at the 30nt cassette. The analysis of the splicing patterns was performed with a RT-PCR strategy based on Agilent technologies. The measurements were taken after 24h of continuous treatment and after 72h of glutamate wash-out. Data represent mean values and standard errors obtained from at least three independent evaluations. Bonferroni correction was used after two-way ANOVA (*p<0,05; **p<0,01; ***p<0,001). After the glutamate treatment, the ADAR2 splicing pattern of the canonical (-47) and alternative form (+47) changed (Fig. S1-A). Two-way ANOVA showed an impact of treatment (p<0.001). The glutamate treatment increased -47nt ADAR2 isoform by 11.1% after 24hr (p < 0.01) and by 13.8% (p < 0.001) after washout. By investigating the -30nt ADAR2 splicing isoform (Fig. S1-B), we also analyzed the splicing pattern involving the deaminase domain. Again, the chronic glutamate treatment led to an increase in short 30nt splicing isoform. Two-way ANOVA showed an effect of treatment (p < 0.001), time (p < 0.001) and time/treatment interaction (p < 0.01). After 24hr of treatment the short isoform increased by 26.6% (p < 0.001) and 38.4% (p < 0.001) after 72hr of glutamate washout. Fig. S3: Glutamate treatment increased Calpain-dependent αII-spectrin breakdown. Representative immunoblots of control (ctr) and 24h glutamate treated (Glu) samples. After the glutamate treatment, we observed an increase in Calpain-dependent αIIspectrin cleaved fragments (145-150kDa), indicating the activation of Calpain3-4 * Fig. S4: Expression level of RPS14, hnRNPA1B2, SRSF9 and SHMF1 mRNA by qPCR after 24h of treatment with the Glu and BAPTA/am (Ca2+ chelator), Calpain inhibitor and KN93 (CAmKII inhibitor) treatment. Data are reported as Log2 of the expression ratio R=2ΔΔCt (expression levels of control samples are equal to 0) and represent mean values and standard errors obtained from at least three independent evaluations. Bonferroni correction was used after one-way ANOVA (*p<0,05; **p<0,01; ***p<0,001). Glutamate treatment increased the expression level of SRSF9, while inhibition of Ca2+ entry, CamKII activity and Calpain activity did not alter the expression levels of the four ADAR cofactors. 1. Barbon A, Fumagalli F, Caracciolo L, Madaschi L, Lesma E, Mora C, et al. Acute spinal cord injury persistently reduces R/G RNA editing of AMPA receptors. J Neurochem 2010; 114:397-407. 2. Gottwald E, Muller O, Polten A. Semiquantitative reverse transcription-polymerase chain reaction with the Agilent 2100 Bioanalyzer. Electrophoresis 2001; 22:4016-22. 3. Schober ME, Requena DF, Davis LJ, Metzger RR, Bennett KS, Morita D, et al. Alpha II Spectrin breakdown products in immature Sprague Dawley rat hippocampus and cortex after traumatic brain injury. Brain Res 2014; 1574:105-12. 4. Wang KK. Calpain and caspase: can you tell the difference? Trends Neurosci 2000; 23:20-6.