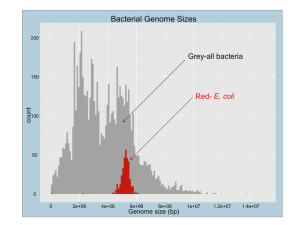
mmi12144-sup-0001-suppl
advertisement

Supplementary Information. Fig. S1. Characterisation of O antigen-producing E. coli K-12 strains. (A) Bacteriophage P1 resistance assay. Bacterial strains were struck perpendicularly across a line of P1 lysate. A zone of clearing was observed for P1 sensitive cells, whilst the resistant strain DFB1655 L9 grew across the lysate. (B) CaCl2 transformation assays. Equivalent amounts of CaCl2 competent E. coli MG1655, DFB1655 L5 and DFB1655 L9 cells were transformed with 0.3 g of pJB42 plasmid as in (3). (C) Bactericidal serum assays. Mid logarithmic phase cultures of each strain were exposed to undiluted human serum and viable bacterial counts were determined after 45, 90 and 180 minutes. Fig. S2. (A) The panel shows the growth of E. coli MG1655, DFB1655 L5 and DFB1655 L9 in LB medium with shaking at 37ºC. (B) The figure shows the effect of triclosan on the growth of MG1655, DFB1655 L5 and DFB1655 L9. Cultures were grown for 90 minutes in LB medium after which triclosan were added as indicated. Triclosan concentrations are in g ml-1. Fig. S3. Western blot analysis of outer membrane proteins. Cultures of strains E. coli MG1655, DFB1655 L5 and DFB1655 L9 were grown to mid logarithmic phase, total protein samples were prepared and equal amounts of protein subjected to Western blotting using anti-OmpF, anti-BamD and anti-RNA polymerase subunit antisera. Note that anti-OmpF antiserum cross reacts with the porins OmpF, OmpC and OmpA (2). Detection of RNA polymaerse subunit is used as a protein loading control. (B) O antigen production does not alter efflux. Hoescht 33342 efflux assays. The figure shows the accumulation of Hoescht 33342 dye within E. coli MG1655, DFB1655 L5 and DFB1655 L9 in the presence and absence of the efflux pump inhibitor (PAβN) after 60 minutes. There was no significant difference between strains under either condition as judged by a student t-test. (C) Viability of E. coli MG1655 and LPS-mutants L5 and L9 during growth. Culture samples from mid-exponential (exp) and stationary (stat) growth phases were stained with PI and BOX viability dyes and analysed by flow cytometry. The fluorescence data is shown as dot plots with green (BOX; FITC) and red (PI; PE-Texas Red) fluorescence intensities plotted on vertical and horizontal axes respectively. The cell populations stain as follows: live cells - no staining, dead cells - PI+BOX-positive, stressed cells (cells that have lost membrane potential) – BOX-positive. Fig. S4. A silver stained SDS PAGE gel of LPS only samples from E. coli 042 based strains. Cultures of strains E. coli 042 (lane 2) and DFB042 (lane 3) were grown to mid logarithmic phase and LPS samples were prepared and subjected to SDS PAGE and silver staining. SDS PAGE markers were loaded in lane 1. Fig S5. O antigen biosynthesis decreases biofilm formation in E. coli K-12. (A) Biofilm formation by strains MG1655, DFB1655 L5 and DFB1655 L9. Cells were grown overnight in flat bottomed polystyrene microtitre plates, containing LB medium. Biofilm formation was quantified using crystal violet staining. There was a significant difference between MG1655 and DFB1655 L9 as judged by the student t-test (P<0.001). (B) Fluorescence microscopy images of biofilms formed by MG1655, DFB1655 L5 and DFB1655 L9 cells. All strains carried the GFP-expressing plasmid pJB42. Bacteria were grown in flow chambers, containing LB medium, and the ability of each strain to adhere to the glass surface was examined using fluorescence microscopy after 24 and 42 h of growth. Fig. S6. O antigen does not alter lysozyme resistance. Cultures were grown for two hours in either the (A) absence or presence of (B) 1 mg ml-1 or (C) 2 mg ml-1 lysozyme before calculating CFU ml-1. No difference was detected. In contrast adding (D) 1mg ml-1 lysozyme with 10 mM EDTA led to a large drop in CFU ml-1 in all strains. Table S1. Chemical compounds which effected the growth of DFB1655 L9 and MG1655 differently during a Biolog Phenotypic MicroArray analysis. Biolog Compound Difference a Mode of action b Phenotypes Gained: Tolylfluanid Trifluorothymidine 129 187 Fungicide, phenylsulphamide Nucleic acid analog, pyrimidine, DNA synthesis -252 -198 Lipid synthesis, fatty acid inhibitor Membrane, detergent, cationic Phenotypes Lost: Triclosan Poly-L-lysine a Differences between the strains are shown in arbitrary units. The average signal for each PM array well was calculated as the mean of the signal from DFB1655 L9 or MG1655 in two independent PM array experiments. The metabolic differences between the strains are shown as the arithmetic difference of the mean DFB1655 L9 signal minus the mean of the MG1655 signal for each test well. Wells where there was no difference are not shown. Wells in which there was a greater signal from DFB1655 L9 than MG1655 are shown as positive values (phenotypes gained) whilst lower signals are shown as negative values (phenotypes lost). b Note that as strain DFB1655 L9 carries pJP5603/ wbbL, cells are resistant kanamycin and as a consequence cells were also found to be resistant to related aminoglycoside antibiotics Table S2. Strains, plasmids and primers used in this work. Bacterial strains. Relevant genotype. Reference. MG1655 K-12 strain. F- - rfb-50 rph-1 (CGSC7740) (1) DFB1655 L9 MG1655 with pJP5603/ wbbL integrated into the This work rfb gene cluster. O antigen producing strain. DFB1655 L5 MG1655 with pJP5603/ wbbL integrated into the This work rfb gene cluster. O antigen producing strain. WG1 DH5 pir K-12 strain. F+ rfb-51 F1-1(CGSC5073) (2) recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 (3) deoR (lacZYA-argF)U169 pir+ S17-1 pir pro res mod+ RP4-2 Tet::Mu-Kan::Tn7 (4) 042 Wild type pathogenic enteroaggregative E. coli (5) DFB042 E. coli 042 with pCVD442/ wbaC integrated into This work the rfb gene cluster. Rough LPS strain. OP50 E. coli B strain (6) Bacterial plasmids. pET20b T7 expression vector (AmpR) Novagen pET20b/wbbL pET20b carrying wbbL This work pET20b/wbbL1 pET20b carrying wbbL, which contains a 1 bp This work deletion of the 10th nucleotide of wbbL. pJP5603 3.1 kb R6K-based suicide vector (KanR) pJP5603/ wbbL pJP5603 carrying wbbL. pCVD442 kb R6K-based suicide vector (AmpR) pCVD442/wbaC pCVD442 carrying an internal fragment of wbaC. (7) This work (8) This work pDOC-C Gene doctoring plasmid (AmpR) (9) pJB42 GFP expressing vector constructed from pDOC-C This work Primers (all are shown 5' to 3'). WbbL(NdeI) CCCGAATTCATATGGTATATATAATAATCGTTTCCC WbbL(HindIII) CCCAAGCTTCTCGAGTTACGGGTGAAAAACTGATGAAATTC WbbLFW GTATTAGTTGGGTTTGATGAACTAG M40Rev CAGGAAACAGCTATGAC wbaCUP CCCCTCTAGACTTGTGCAAAAGTTAAGAGAC wbaCDWN CCCCTCTAGATTCCATACCCCTCGGCAAGCGAATG References for Supplementary Information. 1. Blattner FR et al. (1997) The complete genome sequence of Escherichia coli K12. Science 277:1453-1462. 2. Bachmann BJ (1990) Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev 54:130-97. 3. Elliott SJ, Kaper JB (1997) Role of type 1 fimbriae in EPEC infections. Microb Pathog 23:113-118. 4. Simon R, Priefer U, Pohler A (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. 5. Nataro JP, et al. (1995) Heterogeneity of enteroaggregative Escherichia coli virulence demonstrated in volunteers. J Infect Dis 171(2):465-468. 6. Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77:71-94. 7. Penfold RJ, Pemberton JM (1992) An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118:145-146. 8. Mobley HL, et al. (1993) Isogenic P-fimbrial deletion mutants of pyelonephritogenic Escherichia coli: the role of alpha Gal(1-4) beta Gal binding in virulence of a wild-type strain. Mol Microbiol 10:143-155. 9. Lee DJ et al. (2009) Gene doctoring: a method for recombineering in laboratory and pathogenic Escherichia coli strains. BMC Microbiol 9:252. 10. Liu D, Reeves PR (1994) Escherichia coli K12 regains its O antigen. Microbiology 140:49-57. 11. Bennion D, Charlson ES, Coon E, Misra R (2010) Dissection of β-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol Microbiol 77:1153-1171. 12. Sambrook J, Russell DW (2001) Molecular Cloning: A laboratory manual, 3 ed, vol. 1-3. Cold Spring Harbor Laboratory Press, Cold Spring harbor, New York.