Muscle Cell Composition
advertisement

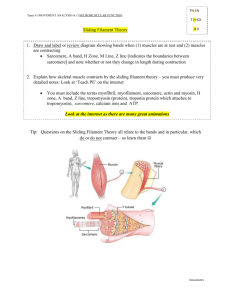
Module 2: Muscle Cell Composition and Structure Introduction Muscle contraction is one of the most studied forms of locomotion in animals. Given the fact that this form of locomotion is specific to animals, muscle is considered a late evolutionary development. Muscle contraction, for the purposes of locomotion, is carried out by skeletal muscle. If we look closer at the structure of the skeletal muscle, we find that the fibers are actually cells that are multinucleated. These cells form during muscle development by the fusion of muscle cell precursors called myoblasts. Although, each muscle cell can have many nuclei, much of the volume of the cytoplasm is composed of myofibrils. A myofibril is a long cylindrical structure of about 1-2 μm in diameter. If we look more closely at the myofibril, we see a repeating set of mini-contractile units known as sarcomeres, which are the most basic contractile units in the cell. The sarcomere is composed of several important proteins that allow for contraction. For instance, the thick filaments in the sarcomere are composed of myosin motors. The myosin motors are actually composed of two different proteins, the myosin light chain, and the myosin heavy chain. Under the microscope, these thick filaments appear black. Furthermore, if we look at the thin filaments, they are composed of actin, and appear white underneath the microscope. These three proteins just described are not the only ones highly expressed in muscle cells. A number of other proteins are highly expressed and are required to properly bundle the myosin into thick filaments, and to bundle the actin into thin filaments. To view all highly expressed muscle cell proteins see chart on the next page. Protein Titin Dystrophin Filamin Myosin Heavy Chain Spectrin M1/M2 M3 C Protein Nebulin α-actinin Gelsolin Fimbrin Actin Tropomyosin Troponin T Myosin Light Chain Troponin I Troponin C Thymosin Size (kD) 3000 400 270 210 265 190 150 140 107 100 90 68 42 35 30 25 19 17 5 *adapted from Bio-rad literature Each myosin molecule contains a head that is bound to ATP. In order for the contraction to occur, the myosin head binds the actin. Then, ATP is hydrolyzed allowing the slide along the actin filament, thus shortening the sarcomere. In order for the muscle to relax, the opposite must occur and the sarcomere must again become elongated. Given the fact that these highly expressed proteins are important for muscle cell structure, we will investigate the results from our acrylamide gels from module 1, and we will then move on to do some deeper study of these proteins through bioinformatics. Procedures: 1. Obtain your stained gel from last week and take a picture on the light box. Be sure to have a ruler next to it. 2. Place your gel in a ziplock bag with a small amount of distilled water. This will allow you to store your gel if you need to take another picture if you need to. 3. Create an Excel spreadsheet, calculate Rf values for your Kaleidoscope protein standards. 3. Prepare a standard curve using the molecular weights of the protein standards and the Rf values that you determine. 4. Create another spreadsheet for your fish samples. Choose 1 of your lanes, and calculate the Rf values for each of the bands you see. 5. Calculate the molecular weights of these proteins using your standard curve. 6. Use the molecular weights from step 5 and the chart above to identify the proteins in your fish sample 7. Determine the molecular weights and identify the proteins for each of your other samples. Add these to your spreadsheet from step 4. Note if there are certain proteins missing from each sample. 8. View your protein profiles on the gel. Determine which are more similar to each other and which are different. The fish with similar profiles that are more evolutionarily related, and than those with different profiles. Bio-informatics: 1. Choose one of the three critical proteins that compose a sarcomere (see introductory materials). Go to the protein database from pubmed.org. Type in that protein name, along with the genus/species name. 2. Click the link to the protein and copy the sequence into a word file. Be sure you obtain the sequence in FASTA format 3. Take the sequence of the protein you just obtained in step 2. Perform a BLAST search with your sequence. 4. Note the number of homologs you received. Determine whether there are any homologs from any of the other fish samples in your list. If there are homologs, copy their sequences into your word file in the FASTA format. 5. Note the similarities and identities between the homologs you received in step 4. 6. Take your homolog sequences and align them using Clustal W. Note if there are any regions with a high level of identity. These regions are considered highly conserved domains, and are important for the function of the protein. 7. Analyze the cladogram you received from the alignment. Does this cladogram match your pervious conclusions from Part I step 8 with regards to evolutionary relationships. 8. Perform steps 1-6 for the other 2 critical proteins that compose sarcomere structure. 9. Perform steps 1-6 for 2 other proteins from the chart above.