Handout 1 – Values & means
advertisement

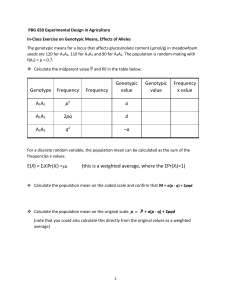
Sanja Franic, VU University Amsterdam 2010 In contrast to classical Mendelian genetics, which deals with the inheritance of interindividual differences in traits along which individuals can be divided into distinct categories (e.g., eye color), quantitative or biometrical genetics is concerned with inheritance of interindividual differences in traits which vary continuously (i.e., quantitative traits, e.g. height). The fact that the intrinsically discontinuous variation in the type of alleles present at the genome may yield continuous variation in observed traits is explained by a) the supposition of polygenic inheritance (quantitative traits are assumed to be affected by genes at multiple genetic loci, whose contribution to the variation in the phenotype is small in comparison to effects of other sources of variation), and b) non-genetic variation, which is truly continuous, being superimposed on the genetic effects on the phenotype. Given the intrinsic reliance of genetic models upon the quantitative genetic theory-based predictions of genetic and environmental covariation between individuals of differing degrees of genetic relatedness, before addressing genetic applications of SEM, we will first review how those predictions are derived. Values and means. The measured value of a trait, or its phenotypic value (P), is typically conceptualized as a sum of two components, one attributable to the particular assemblage of segregating genes relevant to the phenotype in question (the genotypic value, G), and the other to all of the non-genetic factors affecting the phenotype (environmental deviation, E).1 The mean environmental deviation in the population is typically scaled at zero; thus the mean phenotypic value equals the mean genotypic value. The aim of succeeding sections will be to demonstrate the derivation of the average degree of genetic resemblance between relatives; in this light, we focus primarily on the genotypic value. 1 In the text that follows we refer only to the component of P that varies in the population; thus the effects of monomorphic genes, as well as the non-variable aspects of the environment, are ignored (but may be modeled by addition of appropriate constants). 0 Sanja Franic, VU University Amsterdam 2010 Consider, for instance, a single locus with two alleles, A1 and A2. The genotypic values of the two homozygotes (A1A1 and A2A2) and that of the heterozygote (A1A2) may be denoted +a, -a, and d, respectively (Figure 1). The point of zero genotypic value is defined as the midpoint between the two homozygotes. The value of d reflects the degree of genetic dominance: in the absence of dominance, d = 0; if A1 is dominant over A2, d > 0; if A2 is dominant over A1, d < 0; in case of overdominance, d > a or d < -a. 1 Sanja Franic, VU University Amsterdam 2010 Table 2 Derivation of mean genotypic values (G), breeding values (A) and dominance deviations (D) Genotype A1A1 A1A2 A2A2 Genotypic value (gi) a d -a Genotype frequency(fi) p2 2pq q2 Mean genotypic value (μgi = gifi) p2a 2pqd -q2a Parental gametes A1 Mean gen. value across genotypes: μG = ∑μgi = a(p – q) + 2dpq Frequencies of genotypes produced Mean values of genotypes Average effect of allele A1A1 A1A2 produced (μGj) (αj = μGj - μG) p q pa + qd α1 = q[a + d(q – p)] pd – qa α2 = – p[a + d(q – p)] A2 p A2A2 q Average effect of allele substitution: α = α1 - α2 = a + d(q – p) Breeding value (Ai) 2α1 = 2qα α1 + α2 = (q – p)α 2α2 = –2pα E[A] = ∑Aifi = 2p2qα + 2pq(q – p)α – 2pq2α = 2pqα(p + q – p – q) = 0 Genotypic value (Gi = μgi - μG) 2q(α – qd) (q – p)α + 2pqd –2p(α + pd) E[G] = ∑Gifi = 2p2q(α – qd) + 2pq[(q – p)α + 2pqd] – 2pq2(α + pd) = 0 2pqd –2p2d E[D] = ∑Difi = –2p2q2d + 4p2q2d – 2p2q2d = 0 Dominance deviation (Di = Gi – Ai) –2q2d 2 Sanja Franic, VU University Amsterdam 2010 Figure 1. Arbitrarily assigned genotypic values in a system with 2 alleles (Falconer & Mackay, 1996). Table 1 Genotype frequencies in the offspring generation as a function of allele frequencies in the parental generation Paternal gametes and their frequencies Maternal gametes and their frequencies A (p) a (q) A (p) AA (p2) Aa (pq) a (q) Aa (pq) aa (q2) If the relative frequencies with which the A1 and A2 alleles occur in the population of interest are denoted p and q, respectively (p + q = 1), the frequencies of the genotypes arising from the process of random mating2 between individuals within this population are given by the binomial expansion (p + q)2 = p2 + 2pq + q2, as shown in Table 1.3 The mean genotypic value of this locus may be obtained by multiplying the value of each genotype by its frequency and summing over the three genotypes: μG = p2a + 2pqd - q2a = a(p – q)(p + q) + 2dpq = a(p – q) + 2dpq. 2 Mating is random if an individual has an equal chance of mating with any other individual in the population. For effects of non-random mating see e.g. Falconer & Mackay (1996). 3 p2, 2pq, and q2 adequately describe the proportions of genotypes in the offspring generation in populations with no migration, mutation or selection. For effects of migration, mutation and selection see e.g. Falconer & Mackay (1996). 3 Sanja Franic, VU University Amsterdam 2010 The allelic equivalent of the genotypic value is average effect of the allele. The average effect of an allele is the mean deviation from the population mean of individuals who received that allele from one parent, the other allele having come at random from the population (Table 2). For instance, if a number of gametes carrying the A1 allele unite at random with gametes from the population (where p is the frequency of the A1 allele and q of the A2 allele), the frequencies of the genotypes produced will be p of A1A1 and q of A1A2. Taking into account these frequencies and the genotypic values associated with each genotype, the mean genotypic value of the locus may be expressed as pa + qd. Subtracting the population mean from this expression yields the expression for the average effect of the A1 allele: α1 = q[a + d(q – p)]. Correspondingly, the average effect of A2 is α2 = – p[a + d(q – p)]. The average effect may also be expressed in terms of the average effect of gene substitution, which is simply the difference between the average effects of the two alleles: α = α1 – α2 = a + d(q – p). The average effects of the two alleles may be conveyed in terms of the average effect of gene substitution: α1 = qα, and α2 = -pα (Table 2). Summing the average effects over the two alleles at a locus yields a component of the genotypic value termed the breeding value or the additive genotype (A). The remainder of the genotypic value is the dominance deviation (D). Dominance deviation arises in the presence of genetic dominance (i.e., within-locus interaction between alleles), and reflects the possible non-additive effects arising from the allelic pairing at a locus. It may be derived as D = G – A (Table 2). If the genotypic value refers to an aggregate value of genotypes across more than one locus, the expression for G takes on the form: G = A + D + I, where I stands for interaction deviation4 arising from possible non-additive gene co-action across loci (i.e., epistasis). 4 Alleles may interact in pairs or threes or higher numbers, in expression (x.x) aggregate interactions of all sorts are treated together as a single interaction deviation. 4