Appendix Supplementary Table 1Strains, plasmids, and primers
advertisement

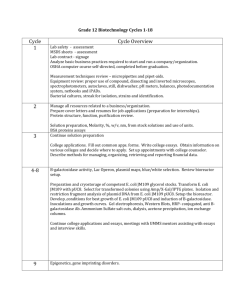
1 Appendix 2 Supplementary Table 1Strains, plasmids, and primers used in this study Description Reference E. coli strains JM109 recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1Δ(lacproAB)/F′[traD36 proAB+lacIq lacZΔM15] Invitrogen JM109 K8-11 JM109 derived, harboring φ80 attP inserted into lacZ, yhiS, xylA, and malE sites This study EC100Dpir116 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galUgalKλ-rpsLnupGpir-116(DHFR) TransforMaxTM T2 multiple copies of Ppdc-yfjBintegrated into JM109 This study Plasmids pAH57 pSC101 replicon, Xisλ and Intλ, AmpR (Haldimann and Wanner 2001) pAH123 pSC101 replicon, Intφ80, AmpR (Haldimann and Wanner 2001) pattP80 R6Kγreplicon, φ80attP, KanR This study pE76 Ppdcpromotercontrolled yfjB gene, AmpR (Li et al. 2009) p80T12 Two copies of Ppdc-yfjB inserted into pattP80, KanR This study pBHR68-gfp gfp inserted into pBHR68, AmpR This study Primers T1F 5’-GAAACACGGAGTGTCTAGATTACGCTCATGATCGC T1R 5’-GAAAAAGCTTATTGCTAGCAGTAAAGAGGCTGG T2F 5’-GAAAAAAGCTTTCTAGATTACGCTCATGATCGC T2R 5’-GAAAAGCCACATAGGCATTGCTAGCAGTAAAGAGGCTG 3 4 All oligonucleotides were synthesized by AuGCTBiotechnology (Beijing, China). Restriction 5 endonuclease digestion sites were underlined. 1 1 Supplementary Table 2qPCR primers used for transcriptional analysis Primers Sequence (5’-) phaC_F AAGTCCCAACCATTCAAG phaC_R AGAAGTCCTTCATGTAGC phaA_F ATCAAGAGCTATGCCAAC phaA_R GTTCACATTGACCTTGGA phaB_F GAGGTTGATGTGCTGATC phaB_R GACGAGATGTTGACGATG yfjB_F AAAGGTTACGAGGTCATC yfjB_R CGGTTGATTCCAATAACTT pgi_F TGTTCTTGGTAGCATCTAA pgi_R ATGTTGGCAGTATCAATAC gapA_F TACTACCGCTACTCAGAA gapA_R TCAGGTCAACTACAGATAC zwf_F AAGAGCAGCAATACAGAA zwf_R AGATTCAGTTCAGGTGTT gnd_F ACTGGTTCCTTACTATACG gnd_R TCACGATTACGACGAATA icd_F CTGAACGGTGACTACATT icd_R AGAATAATAGAGCCAGGATT mdh_F ATATCATTCGTTCCAACAC mdh_R CTTCAACCACTTCAGTAC pntA_F CGGAAGTGAAAGAACAAG pntA_R CGGTGGTGACAATGATAT pntB_F AAGTTGAAATGACCGAAAT pntB_R GATGAAGATACCGAGGAA udhA_F CGAAGAGTACGAGAAGAT udhA_R CTGATACATGCTGTTGAC 2 1 Supplementary Table 3PCR primers used for genome integration verification Primers Integration site K8_F Sequence (5’-) CAGTGAGCGCAACGCAATTA lacZ K8_R GAAATACGGGCAGACATGGC K9_F TGGGGCGTGACATGAATATC yhiS K9_R CAGGGACTTGTTCGCACCTT K10_F AGCGAGCGCACACTTGTGAA xylA K10_R TATCGCTACCGATAACCGGG K11_F GGGAGGATGAGAACACGGCT malE K11_R AGCACTGACCATTTCAGCGC 3 1 Supplementary Fig.1PCR verification of NAD kinase integration 2 3 PCR primers used for genome integration verification were listed in Supplementary Table 3. 4 E. coli T2 was inoculated into Luria-Bertani medium and maintained by transferring into 5 fresh medium every 12 h for 7 days. (A), original culture; (B), 7-day culture. 4 1 2 Supplementary Fig.2Expression of gfp gene under the control of phaCAB promoter in E. coli JM109 and T2 3 4 5 6 7 E. coli JM109 and T2strains harboringpBHR68-gfp were cultivated in mineral saltsmedium at 37 °C for 48 h,respectively. Value 100 corresponds to equal gene expression relativeto JM109 and values above 100 correspond to foldincrease. Data were the means of triplicate experiments. 8 9 5