introduction - NE-CAT
advertisement

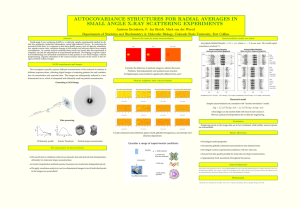
SAXS – its Genesis and Capacity to Meet the Challenges for Structural Systems Biology Robert P. Ramboa, Greg L. Huraa,, Michal Hammela, John A. Tainera,b a Lawrence Berkeley National Laboratory, Berkeley, CA; bDepartment of Molecular Biology & The Skaggs Institute for Chemical Biology, The Scripps Research Institute, La Jolla, CA The doubling of DNA sequencing output every 9 months outpaces advances in computer storage and performance. We propose this fast marks the switch from the computer age to the age of biology; it offers the potential to harness 3 billion years of evolution to solve problmes in medicine, technology, and climate change. However information on the conformations and assemblies of proteins, DNA, and RNA, plus their detailed structural chemistry will be needed if we are to harness sequence information to define and control outcomes in biology. We are developing Small Angle X-ray Scattering (SAXS) combined with crystallography as a premiere tool for defining macromolecular conformations and connections suitable to join proteins to pathways at the systems biology scale1-4. Yet, many misconceptions exist about SAXS. The perception has been that SAXS fails as a robust structural technique due to limited resolution. This is not correct. This talk will examine the genesis and promise of SAXS for efficient and robust modeling of solution structures. SAXS was initially used to measure a physical property of the particle, i.e. Rg. So experimental results were compared to ultracentrifugation data. SAXS has been under appreciated as a legitimate structural technique for biological samples. Why? The original limitations of SAXS as a technique to determine solution structures came primarily from three problems that can now be overcome: 1) difficult data collection, 2) difficult interpretation, and 3) insufficient data sampling. Curent limitations on SAXS do not primarily come from limited resolution, but rather the quality of the resulting models. Advances in SAXS are making this technique powerful and robust for efficiently examining complexes in solution, as aided by computer interfaces allowing biologists to do these experiments4. Structures of flexible systems and complexes that switch from one conformation to another can now be distinguished and support the promise of SAXS for examining the assemblies and conformations of dynamic complexes in solution5-6. In principle, SAXS can provide reliable solution data on small and large macromolecules1-2. In practice, SAXS can be limited by problems in samples and analyses, which can be reduced or avoided3. Emerging results on dynamic complexes show that SAXS has great potential to provide accurate shapes, conformations, and assembly states in solution and inform biological functions in fundamental ways1-6. 1. Putnam, C.D., Hammel, M., Hura, G. L., and Tainer, J. A. (2007) X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution, Quarterly Reviews in Biophysics 40, 191-285. 2. Hura, G.L., Menon, A.L., Hammel, M., Rambo, R.P., Poole, F.L., Tsutakawa, S.E., Jenney, F.E., Frankel, K.A., Hopkins, R.C., Scott, J.O., Dillard, B.D., Classen, S., Adams, M.W.W. and Tainer, J.A. (2009) Rapid and robust proteomics-scale solution structural analyses determined efficiently by x-ray scattering (SAXS) Nature Methods 6, 606-612. 3. Rambo RP, Tainer JA. (2010) Improving small-angle X-ray scattering data for structural analyses of the RNA world. RNA 16, 638-646. 4. Classen S, Rodic I, Holton J, Hura GL, Hammel M, Tainer JA. (2010) Software for the highthroughput collection of SAXS data using an enhanced Blu-Ice/DCS control system. J Synchrotron Radiat. 17, 774-81. 5. Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, Rambo RP, Hura GL, Pelikan M, So S, Abolfath RM, Chen DJ, Lees-Miller SP, & Tainer JA. (2010) Ku and DNA-dependent protein kinase (DNA-PK) dynamic conformations and assembly regulate DNA binding and the initial nonhomollogous end joining complex. J Biol Chem. 285, 1414-1423. 6. Rambo RP, Tainer JA. (2011) Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers 95, 559-571. 1