Tips for protein solution SAXS
advertisement

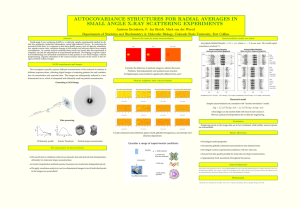
Tips for conducting protein solution SAXS at NSRRC BL23A SWAXS endstation U-Ser Jeng, Chun-Jen Su, and Yu-Shan Huang May 2013 1. Each sample loading requires ~60 l for 2.5-mm cell (12-15 keV), or 30 l for 1.2 mm cell (8-10 keV). For general cases, 2.5 mm with 14-15 keV is suggested. 2. Overseas samples should be shipped in liquid in tubes and transported in 4 C with ice pack. 3. Protein sample concentration can be 1-5 mg mL-1; for high quality high-Q data (0.3-0.5Å-1) 5-10 mg mL-1 is suggested. 4. Exact buffer solution is needed for SAXS background subtraction, “exactly” the same (or as close as possible) as that used for the corresponding protein solutions. Corresponding dialysis remainder for each concentrated sample solution is the ideal buffer solution for scattering background measurement. 5. Calculate zero angle scattering intensity I(0) value of the protein solution, using the provided 23A Io-Excel spreadsheet for an estimate of an appropriate protein concentration for SAXS. Choose a sample concentration for an I(0) value reasonably higher than 0.0167 cm-1 (of water scattering intensity). 6. Prepare samples of different concentrations (such 1-2-4 mg/ml) for checking inter-particle interactions. 7. Prepare sample solutions of different concentrations from a concentrating process (and meanwhile get the corresponding buffer solutions); over-concentrated sample solution may result in sample aggregation that may not be rescued by further dilution, 8. Aggregated samples may be rescued by centrifuge refrigerator or off-line FPLC at the E171 lab by the 23A SWAXS endstation. 9. Conduct size exclusion chromatography if samples are originally purify by ion exchange method; native PAGE is recommended. 10. Conduct HPLC/FPLC and dynamic light scattering before bringing samples for SAXS. 11. Dry, concentrate, and then freeze the sample protein for transportation may lead to unpredictable aggregation effect in redissolving the sample proteins on onsite for SAXS. 12. Using higher X-ray beam energy and the sample rocking mode during SAXS data collection to reduce radiation damage effect 13. Use multi-frame data taking mode to monitor the radiation damage in the typical data collection. 14. In general, the data q-range should cover ~0.02-0.4 Å-1 for protein mass 1~10K; 0.006-0.025 Å for 10-40 kDa; for mass ~ 100 kDa, 0.003 Å-1 resolution will be needed. 15. For general cases, data q-range should cover 0.01 - 0.3 Å-1. 16. Do sample transmission measurement before and after SAXS data collection, and use the averaged samples transmission in background subtraction 17. If negative scattering intensity is encountered, re-do SAXS and transmission for the reloaded sample and/or buffer solutions.