Gene Expression Regulation
advertisement

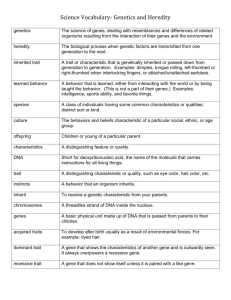
Gene Expression Control Points Ideally, each cell in your body is genetically identical. This means that you have the same two alleles for each of your 30,000 genes in each of your somatic cells. Your cells develop different structures and functions mainly due to the fact that they vary in the combinations of active genes. Recall that the most common purpose of a gene is to build a polypeptide via transcription and translation. A gene is being expressed when a cell works toward this purpose. A gene is being repressed when something interferes with polypeptide production. At any given time, only 3-5% of the DNA in a typical eukaryotic cell is actually expressed. The fact that genes may be expressed (turned on) and repressed (turned off) further complicates genetics. An organism may possess a certain genotype, but if the gene is repressed, it will not exhibit the predicted phenotype. In addition, an organism may be genetically normal, but the inability to express necessary genes may still lead to abnormalities. This packet summarizes how several factors are able to contribute to the expression and repression of eukaryotic genes. Each typed description and most diagrams are taken from your text. In order to improve your chances on your eventual quiz, you’ll want to put the descriptions into your own words in the space provided. Be sure to focus on how the genetic process is expressed or repressed. Chromatin Modification (heterochromatin vs. euchromatin) Euchromatin Nucleosome (cluster of histones) Heterochromatin During interphase, portions of certain chromosomes in some cells exist in the highly condensed state represented in the figure above. This type of interphase chromatin, which is visible with a light microscope, is called heterochromatin, to distinguish it from the less compacted euchromatin ("true chromatin"). What is the function of this selective condensation in interphase cells? The formation of heterochromatin may be a sort of coarse adjustment in the control of gene expression, for heterochromatin DNA is not transcribed. DNA Methylation DNA methylation is the attachment of methyl groups (-CH3) to DNA bases after DNA is synthesized. The DNA of most plants and animals has methylated bases, usually cytosine. About 5% of the cytosine bases in methylated eukaryotic DNA have methyl groups. Inactive DNA, such as that of inactivated mammalian X chromosomes, is generally highly methylated compared to DNA that is actively transcribed, although there are exceptions. Comparison of the same genes in different types of tissues shows that the genes are usually more heavily methylated in cells where they are not expressed. In addition, demethylating certain inactive genes (removing their extra methyl groups) turns them on. Histone Acetylation There is mounting evidence that histone acetylation and deacetylation play a direct role in the regulation of gene transcription. Histone acetylation is the attachment of acetyl groups (-COCH3) to certain amino acids of histone proteins; deacetylation is the removal of acetyl groups. When the histones of a nucleosome are acetylated, they change shape so that they grip the DNA less tightly. As a result, RNA polymerase has easier access to genes in the acetylated region. In other words, histone acetylation and the initiation of gene transcription seem to be coupled structurally as well as functionally. Alternative Splicing The RNA transcripts of some genes can be spliced in more than one way, generating different mRNA molecules. Notice in this example that one mRNA molecule has ended up with the “C” exon and the other with the “D” exon. With alternative splicing, an organism can get more than one type of polypeptide from a single gene. A B C D E A B C D E A B C E A B D E Protein Degradation (proteasomes) A proteasome is an enormous protein complex that chops up unneeded proteins in the cell. In most cases, the proteins attacked by a proteasome have been tagged with short chains of ubiquitin, a small protein . A proteasome recognizes the ubiquitinated protein, unfolds it, and sequesters it in its central cavity . Enzymatic components of the proteasome cut the protein into small peptides , which can be further degraded by cytosolic enzymes. Repressible Operon Circle the correct boldfaced terms. By default… Repressor active/inactive Operon is on/off Operon may be turned on/off when reactant/product is present/absent. Inducible Operon Circle the correct boldfaced terms. By default… Repressor active/inactive Operon is on/off Operon may be turned on/off when reactant/product is present/absent.