Biomolecular Structures and Modeling
advertisement

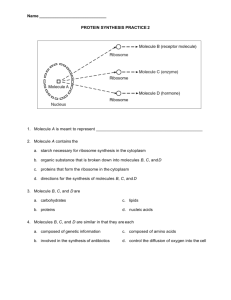
Teaching Notes Monitoring Student Learning To assess if your students have learned about a biomolecular structures and modeling, you can assign relevant Molecule of the Month features for them to read and then answer questions about. Feel free to include additional questions to the basic question set to customize your assessment. Any Molecule of the Month article related to your/your students’ interests may be assigned. For sample responses to the Questions asked about the Molecule of the Month articles - see below. Example molecules include: 1. G-Proteins 2. TATA-Binding Protein 3. Glycolysis 4. Vitamin D Developed as part of the RCSB Collaborative Curriculum Development Program 2015 Teaching Notes 1. Questions Based on the RCSB PDB Molecule of the Month Go to the Molecule of the Month Article at the RCSB PPDB or PDB-101 websites (www.rcsb.org or http://pdb101.rcsb.org/ respectively). Read the article and respond to the following questions. *If there are 3 or more different proteins discussed in the article—e.g. as in the case of the Glycolytic Enzymes—choose any one of the molecules to answer the starred (*) questions. 1. Which Molecule of the Month article did you read? G Proteins 2. About the Featured Molecule(s) a. Function: What is the main biological function discussed in the article? G proteins are molecular switches, associated with various types of membrane receptor molecules, on the inner surface of the cell membrane. Once the receptor binds to its ligand, a message is received by the G proteins, which in turn initiate the signal transduction pathway. b. *Players: Name the key molecule(s) (proteins, nucleic acid, etc.) performing the function(s) listed above? Are there any other molecules mentioned in the article that interact with the molecule being studied – either facilitating or regulating the discussed function? Name the molecule(s). The alpha subunit (with bound GTP) of the G protein performs the main function. Adenylyl cyclase is a molecule that supports this function. When the alpha subunit of the G protein falls off, it moves along the membrane to find adenylyl cyclase and activates it to produce cyclic AMP. c. Big picture: Describe in 3-4 sentences how reading this article helps you understand the function of a living organism or the world around you? G proteins play a huge role in helping our body function efficiently. While they are responsible for carrying important messages and facilitating metabolism, vision, transmission of nerve signals, they are also a key target for many toxins and drugs. Studying the structure of G proteins helps us understand its functions and develop suitable drugs and inhibitors for specific instances of this class of molecules. 3. *Explore the structure-function relationship of the molecule(s) discussed in the article a. Overview: Describe how the shape, size and interaction of relevant molecules discussed in the article help in performing the function Three different protein chains, called alpha, beta and gamma make up the GProtein. The alpha chain undergoes conformational changes upon binding to GTP and GDP resulting in changes in the way they interact with the beta and gamma chains. GTP is larger than GDP by one whole phosphate group. Exchange of GDP to GTP leads to conformational changes, ultimately activating the G protein to interact with the enzyme, Adenylate cyclase. b. Details: Go to “Exploring the Structure” section in the article and analyze the structures shown in detail. i. Basic level: Examine the static images, and JSmol interactive views, where available. If only static images are available, take a screen shot of the image, include it in your answer and explain in 1-2 sentences the structural and functional detail highlighted in it. In articles where the JSmol interactive Developed as part of the RCSB Collaborative Curriculum Development Program 2015 Teaching Notes views are available, take screenshots of at least two different views, and include them in your answer. Explain in 1-2 sentences the structural and functional details that these images highlight. In the GDP bound form all three chains of the G-Protein non-covalently interact with each other (figure labeled GDP (inactive), shown below. Once the larger GTP replaces the GDP, a conformational change occurs, allowing the protein to be in active form (shown below). When the G Protein is in the active form (bottom left side), a short loop on the surface of the protein created by the last phosphate group in the molecule allows it to latch onto the surface, enabling it to later latch on to adenylyl cyclase when found, thus beginning the cyclic AMP cascade. ii. Advanced level: Click on any one of the 4-character accession code (PDB ID) discussed in the “Exploring the Structure” section of the article. This should lead you to the Structure Summary page of that PDB structure. Click on the “3D View” tab and open a JSmol image that can be manipulated. Select from the various visualization/customization options available. Take screenshots of at least two different views highlighting structural details that are important for its function, and include them in your answer. Include the Developed as part of the RCSB Collaborative Curriculum Development Program 2015 Teaching Notes PDB ID that you use, in your answer. Explain in 1-2 sentences the structural and functional details that each of these images highlight. The structure of active form of the alpha chain of G-Protein is seen in the images below (from PDB entry 1gia). The image below shows two domains with the GTP bound between them (highlighted in yellow). One of the domains consists solely of alpha helices, whereas the other bottom part includes both alpha helices and beta sheets. In the image shown below, the same protein structure is shown in space-fill mode, with all side-chains etc. shown. You can barely see the GTP here, since it is concealed beneath the rest of the molecule. Developed as part of the RCSB Collaborative Curriculum Development Program 2015 Teaching Notes 2. Questions Based on the RCSB PDB Molecule of the Month Go to the Molecule of the Month Article at the RCSB PPDB or PDB-101 websites (www.rcsb.org or http://pdb101.rcsb.org/ respectively). Read the article and respond to the following questions. *If there are 3 or more different proteins discussed in the article—e.g. as in the case of the Glycolytic Enzymes—choose any one of the molecules to answer the starred (*) questions. 4. Which Molecule of the Month article did you read? TATA-Binding Protein 5. About the Featured Molecule(s) a. Function: What is the main biological function discussed in the article? TBP recognizes and binds to the TATA sequence on the promoter region. b. *Players: Name the key molecule(s) (proteins, nucleic acid, etc.) performing the function(s) listed above? Are there any other molecules mentioned in the article that interact with the molecule being studied – either facilitating or regulating the discussed function? Name the molecule(s). In the images shown in the article the following proteins are shown: -TBP: blue; protein, which will bind to DNA and help initialize transcription -DNA: red; serves as the template, which is copied in transcription -TFIID: green; a transcription factor, which is made up of TBP and other factors -TFIIB: green; binds to the TBP-DNA complex; this new complex can recruit additional transcriptional factors and promote transcription. c. Big picture: Describe in 3-4 sentences how reading this article helps you understand the function of a living organism or the world around you? Transcription is the first step of gene expression, which is essential to protein production, cell differentiation and therefore our growth. TBP binding to DNA is the first step of the transcription process of eukaryotes. Understanding how TBP binds can help us figure out regulation of the transcription process. 6. *Explore the structure-function relationship of the molecule(s) discussed in the article a. Overview: Describe how the shape, size and interaction of relevant molecules discussed in the article help in performing the function TBP is a small protein that is able to grab the TATA sequence. TBP is composed of two symmetrical halves that seem to sit on either side of the DNA strand and bend it sharply. b. Details: Go to “Exploring the Structure” section in the article and analyze the structures shown in detail. i. Basic level: Examine the static images, and JSmol interactive views, where available. If only static images are available, take a screen shot of the image, include it in your answer and explain in 1-2 sentences the structural and functional detail highlighted in it. In articles where the JSmol interactive views are available, take screenshots of at least two different views, and include them in your answer. Explain in 1-2 sentences the structural and functional details that these images highlight. Developed as part of the RCSB Collaborative Curriculum Development Program 2015 Teaching Notes ii. Advanced level: Click on any one of the 4-character accession code (PDB ID) discussed in the “Exploring the Structure” section of the article. This should lead you to the Structure Summary page of that PDB structure. Click on the “3D View” tab and open a JSmol image that can be manipulated. Select from the various visualization/customization options available. Take screenshots of at least two different views highlighting structural details that are important for its function, and include them in your answer. Include the PDB ID that you use, in your answer. Explain in 1-2 sentences the structural and functional details that each of these images highlight. This representation shows TBP bound to DNA, in a space-fill view colored by amino acid. Arginine and lysine are colored in dark blue while DNA is colored in gray. These interactions are what bind TBP to DNA. This second representation, in cartoon style and colored by secondary structure, shows the symmetry of TBP and how the loops “grab” at the DNA strand to bend it. Developed as part of the RCSB Collaborative Curriculum Development Program 2015 Teaching Notes 3. Questions Based on the RCSB PDB Molecule of the Month Go to the Molecule of the Month Article at the RCSB PPDB or PDB-101 websites (www.rcsb.org or http://pdb101.rcsb.org/ respectively). Read the article and respond to the following questions. *If there are 3 or more different proteins discussed in the article—e.g. as in the case of the Glycolytic Enzymes—choose any one of the molecules to answer the starred (*) questions. 7. Which Molecule of the Month article did you read? Glycolytic Enzymes 8. About the Featured Molecule(s) a. Function: What is the main biological function discussed in the article? These enzymes are the “workers” in the process of glycolysis. For example, Hexokinase starts off glycolysis by transferring phosphate from ATP to glucose to form glucose-6-phosphate. b. *Players: Name the key molecule(s) (proteins, nucleic acid, etc.) performing the function(s) listed above? Are there any other molecules mentioned in the article that interact with the molecule being studied – either facilitating or regulating the discussed function? Name the molecule(s). Hexokinase: transfers phosphate ion from ATP to glucose. Thus ATP is needed by hexokinase to start the glycolysis process c. Big picture: Describe in 3-4 sentences how reading this article helps you understand the function of a living organism or the world around you? Glycolysis is a metabolic process that occurs in almost all cells. During glycolysis glucose is partially broken down to provide energy. It is part of both aerobic and anaerobic respiration. The enzymes discussed in this process are what drive the glycolysis process. 9. *Explore the structure-function relationship of the molecule(s) discussed in the article a. Overview: Describe how the shape, size and interaction of relevant molecules discussed in the article help in performing the function Hexokinase is shaped like a clamp so that when ATP and glucose are bound, the hexokinase will close in on the two and protect them from water. b. Details: Go to “Exploring the Structure” section in the article and analyze the structures shown in detail. i. Basic level: Examine the static images, and JSmol interactive views, where available. If only static images are available, take a screen shot of the image, include it in your answer and explain in 1-2 sentences the structural and functional detail highlighted in it. In articles where the JSmol interactive views are available, take screenshots of at least two different views, and include them in your answer. Explain in 1-2 sentences the structural and functional details that these images highlight. ii. Advanced level: Click on any one of the 4-character accession code (PDB ID) discussed in the “Exploring the Structure” section of the article. This should lead you to the Structure Summary page of that PDB structure. Click on the Developed as part of the RCSB Collaborative Curriculum Development Program 2015 Teaching Notes “3D View” tab and open a JSmol image that can be manipulated. Select from the various visualization/customization options available. Take screenshots of at least two different views highlighting structural details that are important for its function, and include them in your answer. Include the PDB ID that you use, in your answer. Explain in 1-2 sentences the structural and functional details that each of these images highlight. Without glucose bound, hexokinase is shaped like a clamp. When glucose binds, hexokinase closes around it and the groove is much smaller. The first image shows Hexokinase with nothing bound to it. The second image shows Hexokinase closing around ATP and glucose. Developed as part of the RCSB Collaborative Curriculum Development Program 2015 Teaching Notes 4. Questions Based on the RCSB PDB Molecule of the Month Go to the Molecule of the Month Article at the RCSB PPDB or PDB-101 websites (www.rcsb.org or http://pdb101.rcsb.org/ respectively). Read the article and respond to the following questions. *If there are 3 or more different proteins discussed in the article—e.g. as in the case of the Glycolytic Enzymes—choose any one of the molecules to answer the starred (*) questions. 10. Which Molecule of the Month article did you read? Vitamin D Receptor 11. About the Featured Molecule(s) a. Function: What is the main biological function discussed in the article? When vitamin D hormone binds to receptors, proteins involved in calcium transport and utilization are produced. b. *Players: Name the key molecule(s) (proteins, nucleic acid, etc.) performing the function(s) listed above? Are there any other molecules mentioned in the article that interact with the molecule being studied – either facilitating or regulating the discussed function? Name the molecule(s). -Vitamin D receptor acts as a communicator between vitamin D and nuclear responses -9-cis retinoic acid receptor (RXR) pairs up with vitamin D receptor to bind to DNA. -Enzymes make Vitamin D into hormone by adding hydroxyl groups to vitamin D -Another special type of vitamin D binding protein takes individual molecules of the hormone to sites of action. c. Big picture: Describe in 3-4 sentences how reading this article helps you understand the function of a living organism or the world around you? Vitamin D regulates levels of calcium and phosphate in the body. It has especially great impact on intestinal cells. A deficiency of vitamin D can lead to diseases such as Rickets, so it is important to learn about how this receptor functions! 12. *Explore the structure-function relationship of the molecule(s) discussed in the article a. Overview: Describe how the shape, size and interaction of relevant molecules discussed in the article help in performing the function There are two domains of vitamin D receptor. One binds to the vitamin D hormone, while the other binds to DNA. Upon binding Vitamin D, the receptor forms a heterodimer with RXR. b. Details: Go to “Exploring the Structure” section in the article and analyze the structures shown in detail. i. Basic level: Examine the static images, and JSmol interactive views, where available. If only static images are available, take a screen shot of the image, include it in your answer and explain in 1-2 sentences the structural and functional detail highlighted in it. In articles where the JSmol interactive views are available, take screenshots of at least two different views, and include them in your answer. Explain in 1-2 sentences the structural and functional details that these images highlight. Developed as part of the RCSB Collaborative Curriculum Development Program 2015 Teaching Notes Native Receptor: Mutant Receptor: The backbone representation with several atoms shown demonstrates how the receptor surrounds the hormones molecules (spacefilled). One option in the JSmol interactive shows the vitamin D receptor (purple spacefill) in its native form while the other is a mutant form (Rickets disease). The Native receptor representation shows how the hormone receptor should actually work. The mutant receptor has a glutamine at position 305 instead of histidine. This residue fits around the hormone molecules differently and less efficiently (making a weaker hydrogen bond). ii. Advanced level: Click on any one of the 4-character accession code (PDB ID) discussed in the “Exploring the Structure” section of the article. This should lead you to the Structure Summary page of that PDB structure. Click on the “3D View” tab and open a JSmol image that can be manipulated. Select from the various visualization/customization options available. Take screenshots of at least two different views highlighting structural details that are important for its function, and include them in your answer. Include the PDB ID that you use, in your answer. Explain in 1-2 sentences the structural and functional details that each of these images highlight. Developed as part of the RCSB Collaborative Curriculum Development Program 2015