Teaching Notes
advertisement

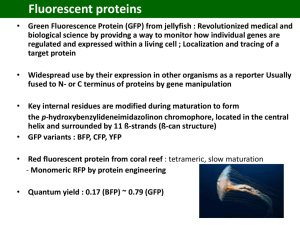
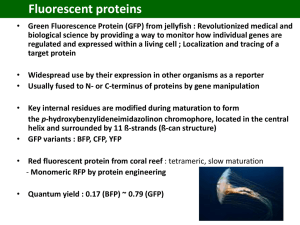
Teaching Notes Make a Paper Model: Green Fluorescent Protein Overview: In this activity you will make a paper model of green fluorescent protein (GFP) structure. Learning Goals: 1. Make a 3D model of transfer green fluorescent protein (GFP) using the template(s) provided. 2. Study the 3D model of GFP to understand its structure, functions and evolution. Educational Standards A. Common Core a. Key Ideas and Details i. RST.11-12.3 b. Integration of Knowledge and Ideas i. RI.11-12.7 B. Next Generation Science Standards a. Practices i. 2. Developing and using models b. Crosscutting Concepts i. 3. Scale, proportion and quantity ii. 4. Systems and system models iii. 6. Structure and function c. Disciplinary Core Ideas i. LS1.A: Structure and Function ii. LS1.D: Information Processing iii. PS1.A: Structure and Properties of Matter iv. PS2.B: Types of Interactions C. Advanced Placement Biology - Essential Knowledge (EK), Learning Objectives (LO), Science Practices (SP) a. EK 1.B.1 i. LO 1.16, SP 6.1 b. EK 4.A.1 i. LO 4.2, SP 1.3 ii. LO 4.3, SP 6.1, 6.4 c. EK 4.A.2 i. LO 4.4, SP 6.4 ii. LO 4.6, SP 1.4 Teaching Notes 1. It is recommended that you read the Molecule of the Month article on Green Fluorescent Protein before attempting to make the paper model. 2. Follow the instructions for making the GFP molecule. 3. Once the model is made, explore and compare it to the JSmol interactives (http://pdb101.rcsb.org/learn/resource/green-fluorescent-protein-gfp-activity-page) showing the experimentally determined models to understand its function and evolution. 4. Click through the various buttons and options for the JSmol interactive model. Developed as part of the RCSB Collaborative Curriculum Development Program 2015 Teaching Notes 5. Note that the proteins GFP and DsRed are distant relatives, with limited sequence identity. The key amino acids essential for cyclization (formation) of the respective chromophores are present at the exact same locations in both structures and play important roles in the protein’s fluorescence. However, the chemistry of the cyclization is slightly different hence the fluorescence color is different. Read more about this in the Molecule of the Month feature on GFP-like Proteins (http://pdb101.rcsb.org/motm/174). 6. The side chains of two highly conserved residues (Glutamate and Arginine) are shown pointing towards the chromophores in the cores of the barrel shaped proteins. The protein core is hydrophobic and it is unusual to have charged residues located buried in the hydrophobic cores. However, these residues have important roles in cyclization of the chromophore, hence are conserved through evolution. 7. Comparison of the GFP and Ds Red structures show how through evolution the betabarrel fold has been conserved for fluorescent proteins. However, the side chains of amino acids facing the outside of the barrel are different in the various proteins requiring various different oligomeric assemblies (tetramer for DsRed and monomer or dimer for GFP). Developed as part of the RCSB Collaborative Curriculum Development Program 2015