jane12357-sup-0001-Suppmat
advertisement
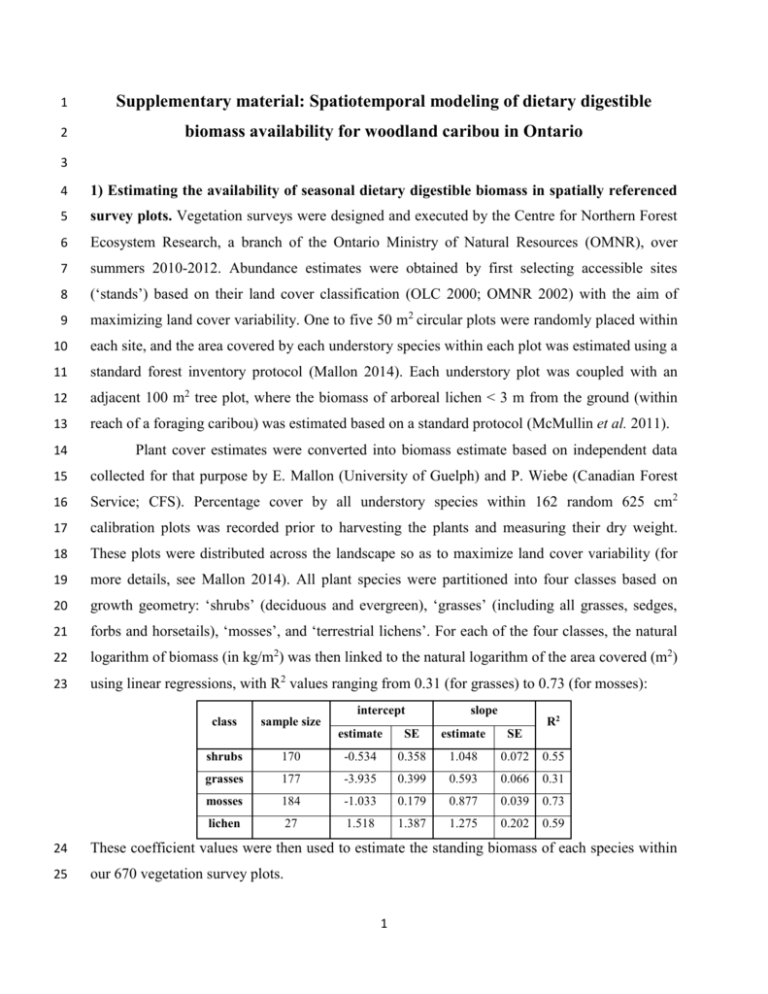
1 Supplementary material: Spatiotemporal modeling of dietary digestible 2 biomass availability for woodland caribou in Ontario 3 4 1) Estimating the availability of seasonal dietary digestible biomass in spatially referenced 5 survey plots. Vegetation surveys were designed and executed by the Centre for Northern Forest 6 Ecosystem Research, a branch of the Ontario Ministry of Natural Resources (OMNR), over 7 summers 2010-2012. Abundance estimates were obtained by first selecting accessible sites 8 (‘stands’) based on their land cover classification (OLC 2000; OMNR 2002) with the aim of 9 maximizing land cover variability. One to five 50 m2 circular plots were randomly placed within 10 each site, and the area covered by each understory species within each plot was estimated using a 11 standard forest inventory protocol (Mallon 2014). Each understory plot was coupled with an 12 adjacent 100 m2 tree plot, where the biomass of arboreal lichen < 3 m from the ground (within 13 reach of a foraging caribou) was estimated based on a standard protocol (McMullin et al. 2011). 14 Plant cover estimates were converted into biomass estimate based on independent data 15 collected for that purpose by E. Mallon (University of Guelph) and P. Wiebe (Canadian Forest 16 Service; CFS). Percentage cover by all understory species within 162 random 625 cm2 17 calibration plots was recorded prior to harvesting the plants and measuring their dry weight. 18 These plots were distributed across the landscape so as to maximize land cover variability (for 19 more details, see Mallon 2014). All plant species were partitioned into four classes based on 20 growth geometry: ‘shrubs’ (deciduous and evergreen), ‘grasses’ (including all grasses, sedges, 21 forbs and horsetails), ‘mosses’, and ‘terrestrial lichens’. For each of the four classes, the natural 22 logarithm of biomass (in kg/m2) was then linked to the natural logarithm of the area covered (m2) 23 using linear regressions, with R2 values ranging from 0.31 (for grasses) to 0.73 (for mosses): intercept class slope R2 sample size estimate SE estimate SE shrubs 170 -0.534 0.358 1.048 0.072 0.55 grasses 177 -3.935 0.399 0.593 0.066 0.31 mosses 184 -1.033 0.179 0.877 0.039 0.73 lichen 27 1.518 1.387 1.275 0.202 0.59 24 These coefficient values were then used to estimate the standing biomass of each species within 25 our 670 vegetation survey plots. 1 26 Next, biomass estimates were converted to digestible biomass estimates. Previously 27 collected biomass samples were used to estimate fractions of Neutral Detergent Fiber (%NDF; 28 including cellulose, hemicellulose and lignin), and Acid Detergent Fiber (%ADF; including only 29 cellulose and lignin), for each species (for methodological details see Mallon 2014). Where 30 possible, fractions were estimated separately for spring (samples taken from May 1st to June 31 15th), summer (June 16th to October 31st) and winter (November 1st to April 31st). Note however 32 that for the majority of species winter samples were never collected and hence summer values 33 were used for both summer and winter. In cases where data at higher taxonomical and/or 34 seasonal resolution were available in the literature, literature values were used (for details see 35 Mallon 2014). Otherwise, missing ADF/NDF values were filled in based on species that were 36 either closely related taxonomically or morphologically similar. To characterize the relationships 37 between ADF, NDF and digestibility, in vitro dry matter digestibility values (IVDMD) over a 38 standard 48 h period were obtained from the published literature on caribou or reindeer for 28 39 plant species together with their respective ADF and NDF values (Mathiesen & Utsi 2000; 40 Storeheier et al. 2002b; a; Ihl & Barboza 2007). Species were again categorized into four classes 41 based on the availability of literature values: shrubs, grasses, mosses and lichens. For the moss 42 group, literature digestibility values (Ihl & Barboza 2007) were coupled with ADF and NDF 43 values from the current study. A linear regression was then fitted within each group to link logit- 44 transformed IVDMD values to logit-transformed ADF, NDF and ΔNDF (the difference between 45 NDF and ADF) values (missing values are due to exclusion in an AIC based model competition): intercept class logit(ADF) logit(NDF) logit(NDF-ADF) sample size estimate SE estimate SE estimate SE estimate SE R2 shrubs 7 -1.295 0.787 -1.647 0.269 - - 0.349 0.433 0.95 grasses 12 -1.865 0.288 -1.228 0.360 - - -1.337 0.515 0.83 mosses 3 1.301 0.568 - - -3.168 0.494 - - 0.95 lichen 6 -1.588 0.494 -0.787 0.178 - - -0.346 0.103 0.84 46 47 In combination with our species-specific ADF and NDF fractions, these coefficient values were 48 then used to estimate the available digestible biomass of each species within 670 vegetation 49 survey plots. 50 Finally, to estimate the dietary digestible biomass and nitrogen available to woodland 51 caribou in each plot, we estimated the fraction of each plant species in the diet of woodland 2 52 caribou. To estimate diet composition, GPS radio-telemetry collars equipped with video cameras 53 (Thompson et al. 2013, 2015) were deployed on 15 female woodland caribou across our study 54 area. After the collars were collected, video files were classified according to time of year and 55 the behavior displayed by the animal. Video clips classified as ‘foraging’ were then watched by 56 field assistants well acquainted with boreal flora, and the number of bites taken from each genus 57 of plants was recorded (for further details see Newmaster et al. 2013 and Thompson et al. 2015). 58 The number of bites per genus was then summed across all animals and normalized to yield diet 59 composition for each of three seasons (spring, summer and winter). For each of our survey plots, 60 we then multiplied the digestible biomass and nitrogen content for each species by its fraction in 61 the caribou’s diet before summing all values to yield the total seasonally available dietary 62 digestible biomass and nitrogen per plot: dietary digestible biomass (kg/m2) dietary nitrogen (kg/m2) minimum 1st quantile median mean 3rd quantile maximum spring 2.60E-05 1.59E-03 3.37E-03 2.69E-02 7.26E-03 8.33E-01 summer 1.60E-05 1.25E-03 2.34E-03 1.94E-02 5.59E-03 5.96E-01 winter 3.00E-06 1.45E-03 3.27E-03 3.42E-02 9.18E-03 1.09E+00 spring 4.38E-06 1.51E-04 3.38E-04 1.03E-03 7.97E-04 2.96E-02 summer 1.72E-06 1.06E-04 1.73E-04 6.36E-04 3.02E-04 1.97E-02 winter 1.86E-07 1.00E-04 2.49E-04 1.00E-03 4.88E-04 3.12E-02 63 64 2) Extrapolation of dietary digestible biomass estimates across the landscape. Our final goal 65 was to generate seasonal landscape projections of forage availability, which requires 66 extrapolation of our forage estimates outside of our survey plots. To achieve this objective, we 67 assembled a set of remote-sensed landscape attributes to serve as potential predictors of forage 68 abundance. MODIS-based Normalized Difference Vegetation Index (NDVI; indicative of live 69 green vegetation) values were obtained via the Land Processes Distributed Active Archive 70 Center at the U.S. Geological Survey Earth Resources Observation and Science Center (temporal 71 resolution: 16 days, spatial resolution: 250 m pixels). To create a temporally-independent 72 seasonal NDVI signature, values were averaged over the four core winter months (December- 73 March; herein ‘winter NDVI’) and the four core summer months (June-September; herein 74 ‘summer NDVI’) from 2010 to 2013. A digital elevation model (ASTER GDEM; spatial 75 resolution: 25 m pixels) was obtained via NASA’s Reverb (http://reverb.echo.nasa.gov/reverb) 76 and used to generate relative elevation maps by subtracting the DEM value at each pixel from the 3 77 average DEM value within a 500 m radius. Land cover classification (spatial resolution: 30 m 78 pixels) was based on the Ontario Provincial Far North Land Cover map (FNLC v1.3.1, ;OMNR 79 2013). This map does not extend south of ~50° N latitude and so was merged with OLC 2000 80 where necessary. Maps included updated disturbances (fire and harvest) for each year of the 81 study based on data provided by (CFS and OMNR personal). 82 Land cover effects were estimated at two spatial resolutions. The land cover class at each 83 survey plot was taken as the dominant class within 30 m radius of the plot location. Hence, each 84 plot was classified as either: ‘coniferous’ (FNLC 18), ‘lowland’ (a combination of FNLC classes 85 6, 9-14), ‘deciduous’ (FNLC 16), ‘mixed’ (FNLC 17), ‘disturbed’ (FNLC classes 19 and 20), or 86 ‘sparse’ (FNLC class 15). However, to avoid singularity, one of these classes had to be omitted 87 from our analysis (here we chose to omit ‘disturbed’). In addition, percent cover for each of 88 seven land cover categories was calculated within a 250 m radius around each of our survey 89 plots. These categories were: ‘coniferous’ (FNLC 18), ‘open lowland’ (FNLC 6, 11 and 13), 90 ‘treed lowland’ (FNLC 9, 10, 12 and 14), ‘deciduous lowland’ (FNLC 8 and 25), ‘deciduous’ 91 (FNLC 16), ‘mixed’ (FNLC 17), or ‘sparse’ (FNLC class 15). To adjust these percent covers to 92 the area of pristine habitat, values were divided by percent cover of undisturbed land (= 1 - 93 (FNLC1 + FNLC2 + FNLC19 + FNLC20)). 94 As the distribution of total available dietary digestible biomass in our samples is strongly 95 right-skewed, values were log-transformed to reduce heteroscedasticity. Mixed effects models 96 were fitted separately for spring, summer and winter using function lme in R, with site (‘stand’) 97 ID as random effects and a Gaussian spatial residual autocorrelation structure within sites. 98 Initially, all potential explanatory variables were included in the model (summer- and winter- 99 NDVI, relative elevation, and land cover at 30 m and 250 m scales) with no interactions other 100 than quadratic terms for NDVI. Function stepAIC from the R package MASS was then used to 101 find the best combination of predictors based on AIC scores. 102 The best models for each season explained 20-30% of the observed variability in the log- 103 transformed response (10-20% variability explained after back-transformation) but tended to 104 undervalue the magnitude of forage availability, particularly at the high end of the forage 105 availability range. We thus calculated a linear correction factor for each model fit as the slope of 106 a zero-intercept mixed effect model of the back-transformed response variable as function of the 107 back-transformed predictions of the models (fixed effects only). 4 log( available dietary digestible biomass ) winter spring summer MLE SE MLE SE MLE SE Intercept -37.89500 8.24236 -34.88061 7.04242 -32.52176 6.67173 con_30m -0.71823 0.27389 -0.71759 0.23475 -0.70305 0.22167 lowland_30m -0.82212 0.28978 -0.52592 0.24842 -0.70441 0.23449 decid_30m -1.10116 0.44055 -0.92677 0.37579 -0.83214 0.35665 decid_250m -1.41566 0.59205 -1.25628 0.50410 -0.98349 0.47931 mixed_250m -0.79759 0.36730 -0.82425 0.31488 -0.69941 0.29726 NDVI.sum 0.85767 0.23159 0.76863 0.19784 0.69684 0.18746 NDVI.sum^2 -0.00647 0.00160 -0.00566 0.00136 -0.00507 0.00129 NDVI.win 0.28505 0.09273 0.23733 0.07937 0.20585 0.07505 NDVI.win^2 -0.00322 0.00127 -0.00277 0.00109 -0.00238 0.00103 correction factor 6.17424 0.82219 5.01664 0.66776 4.98149 0.65295 108 109 These regression coefficients were then used to generate seasonal maps of expected 110 dietary digestible biomass across the entire study area. Areas covered with water and those 111 disturbed the previous year (for winter and spring), or the current year (for summer), were 112 arbitrarily set to zero. 113 5 114 Supplementary material: Spatiotemporal modeling of wolf density 115 116 The procedure described in the main text allowed us to calculate seven population-level 117 wolf density kernels, four for winter (2009-10 to 2012-13; based on a total of 59,050 GPS points 118 from 32 packs), and three for summer (2010-2012; based on 60,196 GPS points from 34 packs). 119 Local density values (in wolves per km2) across these kernels span three orders of magnitude and 120 ranged from 0.0000661 to 0.271939 (with a mean of 0.006813) in summer and from 0.000202 to 121 0.226921 (with a mean of 0.007418) in winter. 122 A regression analysis of local wolf density (log-transformed) as function of local habitat 123 covariates was then performed separately for summer (with a total of 147,901 hexagonal cells, 124 each 0.22 km2 in area) and winter (with a total of 169,738 hexagonal cells). Each hexagonal cell 125 associated with a wolf density value was characterized by temporally-matched values of multiple 126 remote-sensed landscape covariates that were used to predict local wolf density. These included 127 seasonal NDVI, relative elevation, and FNLC v1.3.1 land cover classes (see the above forage 128 modelling section for more details). Cells were also characterized based on their proximity to 129 dumps and settlement (if < 1 km, 1, otherwise, 0), their proximity to roads (if < 500 km, 1, 130 otherwise, 0), and their distance to shoreline of rivers or large lakes. Land cover classes were 131 amalgamated into 9 categories: water (open and turbid classes), open lowland (open fen, open 132 bog, and freshwater marsh classes), treed lowland (treed peatland, treed fen, treed bog, and 133 coniferous swamp classes), deciduous lowland (thicket swamp and deciduous swamp classes), 134 deciduous upland (deciduous treed class), mixed upland (25-75% deciduous, 25-75% coniferous, 135 and mixed treed class), sparse forest (sparse treed class), disturbed (disturbed treed/shrub and 136 disturbed non/sparse classes; representing natural and anthropogenic disturbances), and newly 137 disturbed (< 1 year from fire or forestry disturbance). The coniferous treed class was withheld 138 from the analysis and thus served as a reference class for the resulting inference. 139 Generalized least squares regression models were fitted using function gls in the R 140 package nlme, which allowed explicitly accounting for spatial autocorrelation in the response 141 variable. For each season, model fits were repeated 50 times, each based on a different sample 142 consisting of 2% of the total available data and taken at regular spatial intervals. Averaging the 143 resulting 50 values for each coefficient allowed us to obtain robust coefficient estimates despite 144 the inherent spatial autocorrelation in the data (for further details see Kittle et al. submitted MS). 6 145 Log(wolf density) SUMMER WINTER Average β Average SE Average β Average SE Intercept -7.867193 0.340019 -7.221411 0.229412 le500m -0.000745 0.004297 -0.002081 0.004348 NDVI 0.006325 0.004431 -0.011382 0.002852 NDVI_last -0.001173 0.003000 0.002686 0.002218 dump 0.790474 0.502683 0.872039 0.471592 settle -0.161816 0.543381 -0.080476 0.442302 prime500 0.100127 0.081268 0.024167 0.086373 sec500 0.140625 0.051471 0.046516 0.055456 distshore_km -0.189199 0.050909 -0.111469 0.052324 water 0.121834 0.131908 -0.151999 0.118354 open_low -0.012386 0.161552 -0.431312 0.175744 treed_low -0.029356 0.082822 -0.126191 0.086712 decid_low 0.163702 0.571260 -0.035573 0.592419 lcV16 (decid) 0.163390 0.180313 0.368103 0.182048 lcV17 (mix) 0.095694 0.125011 0.256256 0.133332 lcv15 (sparse) 0.051991 0.291906 -0.043764 0.306415 disturb 0.153453 0.084417 0.060398 0.087992 disturb_new 0.005035 0.297817 -0.155916 0.275190 146 147 The pseudo R2, based on regressing the all observed wolf density values against the averaged 148 model’s predictions, was low (~ 5%), and the linear correlation between predicted and observed 149 was app 0.2. These goodness-fit values indicate a poor model fit and reflect the week links 150 between local wolf density and remote-sensed habitat attributes in our study area. Averaged 151 regression coefficients were then used to project wolf density maps across the entire study area 152 for each season-year combination. 153 7 154 Supplementary material: Spatiotemporal modeling of moose habitat 155 156 Moose occupancy was obtained from fixed-wing aerial surveys flown from Feb. 13 to 157 Mar. 12, 2011 in the extreme northwest of our study area, and Jan. 5 to Mar. 5, 2012 in the 158 extreme southeast of our study area. Both surveys covered roughly 22,500 km2. 1 km transects 159 oriented north-to-south were placed systematically across both landscapes at 5 km intervals to 160 encompass anticipated gradients in disturbance and wolf density. Two observers searched within 161 500 m on either side of the aircraft and recorded the GPS position as well as the number, age, 162 and sex of detected moose. Overall, 4322 km were flown in the northwest survey, with 72 moose 163 sited, and 4236 km were flown in the southeast survey, with 98 moose sited. 164 We identified landscape covariates of biological relevance to moose from land cover, 165 productivity, and weather data. We used FNLC v1.3.1 at a 30 m resolution, modified as 166 described above, to classify forest cover. Because remotely sensed data are inherently error 167 prone, we calculated the difference in mean seasonal NDVI (∆NDVI) for each buffer, estimated 168 as described above, as a correction for available deciduous foliage (i.e., prime moose foraging 169 habitat). We calculated mean snow depth (based on the projection described below) because 170 deeper snow inhibits moose movement and increases vulnerability to predation. Because relative 171 elevation influences dominant vegetation types at a fine scale (i.e., highlands vs. lowlands), we 172 calculated relative elevation within 500 m of a buffer centroid based on digital elevation models 173 from NASA’s Reverb database. We placed circular (250 m radius) buffers around each moose 174 location and characterized landscape covariates within these buffers as proportional coverage for 175 categorical variables (i.e., land cover classes), and as mean values for continuous variables (i.e., 176 snow depth). We created non-overlapping unused buffers spaced at 600 m between centroids 177 along the flight path and similarly calculated landscape covariates within each. In total, we 178 recorded 98 used vs 5997 unused locations in the southeast survey, and 72 used and 6618 unused 179 locations in the northwest survey. 180 We estimated the probability of moose habitat use using logistic regression, per the 181 resource selection function (RSFs) methodology described by Manly et al. (2002). We have 182 excluded deciduous habitat from the model to eliminate perfect collinearity between categorical 183 stand types. Thus selection for stand types represents selection relative to deciduous habitat. To 184 account for any effects of the survey year (the southeast survey was performed one year after the 8 185 northwest surveys), we included the survey identity as a main effect in the RSF (southeast = 0, 186 northwest = 1). We reduced the full model using stepwise AIC and the final model had a 187 Nagelkerke's pseudo R2 of 0.15: MLE SE Intercept -6.97007 0.66988 ∆NDVI 0.11326 0.01375 Relative elevation (m) -- -- Snow depth (m) -0.01362 0.00677 Water -2.95998 0.48004 Lowland -1.90219 0.33182 Mixed -- -- Conifer -2.25297 0.39014 Sparse -3.47724 1.96844 Disturbance -- -- Recent burn -- -- Survey 0.33583 0.18111 188 189 For the purpose of predicting across the entire landscape, the ‘Survey’ effect was 190 averaged (halved) and added to the ‘Intercept’. The resulting logistic model was used to project 191 moose relative probability of use for each 0.22 km2 hexagonal cell across the study area. 9 192 Supplementary Material: Spatiotemporal modeling of snow depth in Ontario 193 194 Observed snow depth data was collected by P. Wiebe (Canadian Forest Service) between 195 2011 and 2013 at several locations across the greater study area, using two methods: fixed 196 cameras that capture daily photos of snow level throughout the winter at the same site, and 197 transects that provides snow depth measurements across different landcover types but with little 198 temporal replication. Combined, these two methods yielded 2326 snow depth observations that 199 could be coupled with spatiotemporal predictors. 200 Each observation was matched with two spatiotemporal predictors; the closest (in space 201 and time) NOAA snow depth value (temporal resolution: 3 hr, spatial resolution: 40 km pixels; 202 NARR dataset DSI-6175), and the closest NDVI value (temporal resolution: 16 days, spatial 203 resolution: 250 m pixels; Land Processes Distributed Active Archive Center). In addition, each 204 sampling location was matched with several spatial covariates. To create a temporally- 205 independent seasonal NDVI signature, values were averaged over the four core winter months 206 (December-March; hereon ‘winter NDVI’) and the four core summer months (June-September; 207 hereon ‘summer NDVI’) for each observation year. A digital elevation model (ASTER GDEM; 208 spatial 209 (http://reverb.echo.nasa.gov/reverb) and used to generate relative elevation maps by subtracting 210 the DEM value at each pixel from the average DEM value within a 500 m radius. Land cover 211 classification (spatial resolution: 30 m pixels) were based on the Ontario Provincial Far North 212 Land Cover Database (FNLC v1.3.1). Each sampling location was classified as either ‘water’ 213 (FNLC 1 or 2), ‘treeless’ (FNLC 6, 11, 13, 21 or 22), ‘deciduous’ (FNLC 8, 25, 16 or 17), 214 ‘sparse’ (FNLC class 9, 10, 12, 14, 15, 19 or 20), ‘coniferous’ (FNLC 18), or ‘road’ (including 215 hydro corridors). resolution: 25 m pixels) was obtained via NASA’s Reverb 216 Model fitting was performed in two stages to appropriately account for the structure of 217 the data set. First, a spatiotemporal mixed effects model was fitted using function lme in R 218 (package nlme) with the natural logarithm of the observed snow depth as the response variable, 219 the natural logarithm of the NOAA snow depth and the NDVI value as the predictors, and the 220 sampling location as random effect. In the second stage, site effects were modeled as function of 221 all spatial covariates (including interactions between seasonal NDVI, relative elevation and 222 landcover), while controlling for sampling method (transect vs. camera), using function gls with 10 223 Gaussian spatial autocorrelation structure. This full model was then reduced based on BIC 224 competition using function stepAIC (package MASS in R). The final model explains 45% in the 225 variation in site effects. Combined, the resulting coefficient estimates explain more than 88% of 226 the observed variability in snow depth measurements across space and time: main effects site effects log( snow depth) MLE SE Intercept 3.20034 0.10079 log(NOAA snow depth + 1) 0.44718 0.01487 NDVI -0.02502 0.00155 Intercept 0.05139 0.02005 Road -0.14147 0.05741 Water -0.35965 0.05381 Relative Elevation * coniferous -0.01877 0.00609 Relative Elevation * deciduous -0.01313 0.00504 Relative Elevation * sparse -0.01693 0.00576 Relative Elevation * winter NDVI 0.00038 0.00010 227 228 11 229 Supplementary material: posterior coefficient estimates 230 caribou ID 1.102 1.112 1.113 1.123 1.130 1.135 1.136 1.141 1.148 1.201 1.202 1.203 2.152 2.155 2.256 2.259 2.261 2.262 2.263 2.268 2.276 2.279 2.280 2.281 2.283 2.284 2.296 2.297 2.313 2.314 θ0 θs 0.011 0.011 0.012 0.011 0.014 0.012 0.014 0.011 0.014 0.009 0.010 0.009 0.014 0.011 0.014 0.010 0.013 0.008 0.016 0.014 0.009 0.011 0.011 0.011 0.013 0.014 0.013 0.010 0.010 0.010 0.000 0.000 0.010 0.008 0.000 0.000 0.000 0.000 0.000 0.006 0.012 0.008 0.000 0.009 0.016 0.000 0.014 0.015 0.000 0.000 0.008 0.000 0.006 0.000 0.017 0.000 0.000 0.017 0.000 0.000 median posterior coefficient values selection α β DDB wolf density moose habitat 0.005 0.000 1.893 -9.612 -0.345 0.047 0.000 2.755 3.276 -39.030 0.055 0.000 -0.408 -17.106 -0.610 0.004 0.000 2.843 -8.324 -4.255 0.085 0.000 1.238 -11.079 -9.583 0.005 0.017 2.263 -2.392 -0.131 0.202 0.000 2.540 -5.149 5.391 0.029 0.000 1.850 -5.420 -3.102 0.161 0.000 -2.642 0.285 -0.246 0.032 0.000 3.619 -5.390 -8.382 0.004 0.000 2.361 -5.236 -0.359 0.002 0.000 1.286 -2.399 -2.041 0.008 0.000 1.949 15.162 -0.933 0.250 0.000 0.915 -14.583 -0.074 0.163 0.000 -3.324 0.085 -4.492 0.016 0.000 6.503 -0.743 -35.609 0.005 0.004 4.552 6.439 -3.561 0.025 0.000 -1.187 -17.931 -3.202 0.009 0.000 2.567 6.952 -10.673 0.037 0.000 2.464 -10.530 -0.456 0.027 0.000 1.465 -16.544 -8.601 0.096 0.000 -1.506 -2.419 -24.347 0.026 0.000 3.301 -8.079 -6.984 0.075 0.000 1.470 -1.376 -8.426 0.040 0.000 0.036 -0.433 -31.024 0.065 0.000 -2.400 -0.184 -9.466 0.009 0.000 3.360 5.592 -0.224 0.019 0.000 0.934 -12.376 -4.826 0.040 0.000 5.050 -7.308 0.221 0.027 0.000 5.821 0.149 -0.225 231 232 12 233 Supplementary material: Bibliography 234 235 Ihl, C. & Barboza, P.S. (2007) Nutritional value of moss for arctic ruminants: a test with muskoxen. Journal of Wildlife Management, 71, 752–758. 236 237 238 239 Kittle, A., Patterson, B., Anderson, M., Moffatt, S., Rodgers, A., Shuter, J., Reid, D., Baker, J., Brown, G., Thompson, I., Street, G., Avgar, T., Hagens, J., Iwachewski, E. & Fryxell, J. Wolves adapt territory size, not pack size to local habitat quality. Journal of Animal Ecology, submitted MS. 240 241 242 Mallon, E.E. (2014) Effects of Disturbance and Landscape Position on Vegetation Structure and Productivity in Ontario Boreal Forests: Implications for Woodland Caribou (Rangifer Tarandus Caribou) Forage. University of Guelph. 243 244 Mathiesen, S.D. & Utsi, T.H.A. (2000) The quality of the forage eaten by Norwegian reindeer on South Georgia in summer. Rangifer, 20, 17–24. 245 246 247 McMullin, R.T., Thompson, I.D., Lacey, B.W. & Newmaster, S.G. (2011) Estimating the biomass of woodland caribou forage lichens. Canadian Journal of Forest Research, 1969, 1961–1969. 248 249 250 251 Newmaster, S.G., Thompson, I.D., Steeves, R.A.D., Rodgers, A.R., Fazekas, A.J., Maloles, J.R., Mcmullin, R.T. & Fryxell, J.M. (2013) Examination of two new technologies to assess the diet of woodland caribou : video recorders attached to collars and DNA barcoding. Canadian Journal of Forest Research, 900, 897–900. 252 OMNR. (2002) Ontario Land Cover Classification. 253 OMNR. (2013) Ontario Provincial Far North Land Cover v1.3.1. 254 255 Storeheier, P., Mathiesen, S., Tyler, N. & Olsen, M. (2002a) Nutritive value of terricolous lichens for reindeer in winter. Rangifer, 34, 247–257. 256 257 258 Storeheier, P., Mathiesen, S., Tyler, N., Schjelderup, I. & Olsen, M. (2002b) Utilization of nitrogen-and mineral-rich vascular forage plants by reindeer in winter. Journal of Agricultural Sciences, 139, 151–160. 259 260 261 Thompson, I.D., Wiebe, P., Mallon, E., Rodgers, A.R., Fryxell, J.M., Baker, J. & Reid, D. (2015) Factors influencing the seasonal diet selection by woodland caribou in boreal forests in Ontario. Canadian Journal of Zoology, in press. 262 13