Homework #3
advertisement
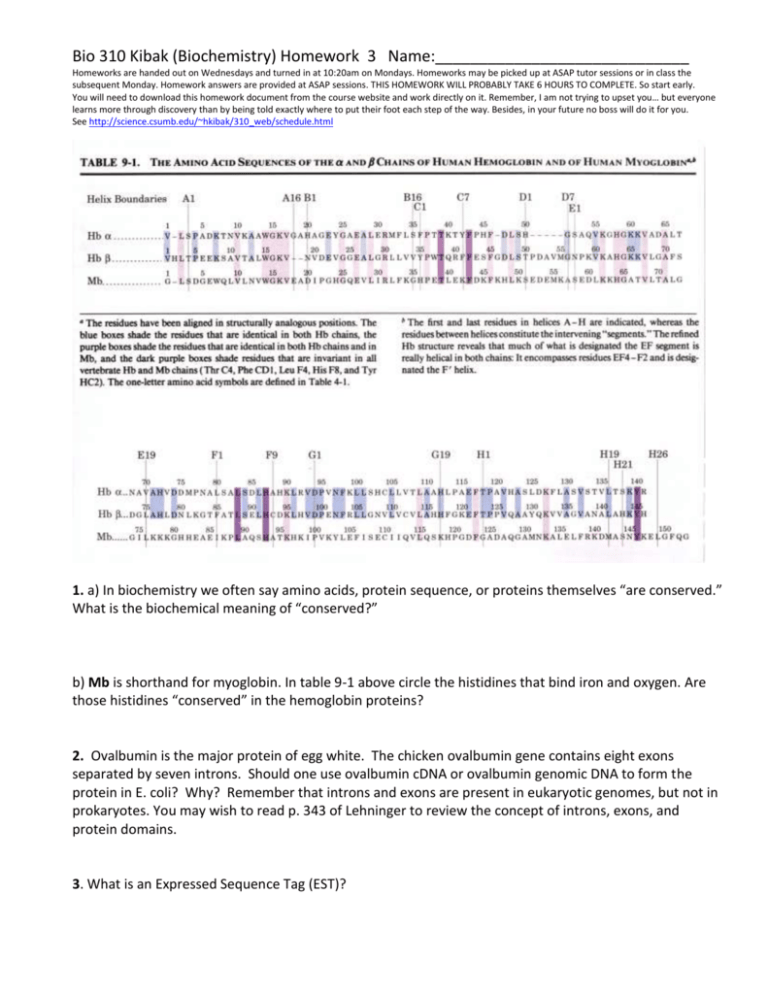
Bio 310 Kibak (Biochemistry) Homework 3 Name:_____________________________ Homeworks are handed out on Wednesdays and turned in at 10:20am on Mondays. Homeworks may be picked up at ASAP tutor sessions or in class the subsequent Monday. Homework answers are provided at ASAP sessions. THIS HOMEWORK WILL PROBABLY TAKE 6 HOURS TO COMPLETE. So start early. You will need to download this homework document from the course website and work directly on it. Remember, I am not trying to upset you… but everyone learns more through discovery than by being told exactly where to put their foot each step of the way. Besides, in your future no boss will do it for you. See http://science.csumb.edu/~hkibak/310_web/schedule.html 1. a) In biochemistry we often say amino acids, protein sequence, or proteins themselves “are conserved.” What is the biochemical meaning of “conserved?” b) Mb is shorthand for myoglobin. In table 9-1 above circle the histidines that bind iron and oxygen. Are those histidines “conserved” in the hemoglobin proteins? 2. Ovalbumin is the major protein of egg white. The chicken ovalbumin gene contains eight exons separated by seven introns. Should one use ovalbumin cDNA or ovalbumin genomic DNA to form the protein in E. coli? Why? Remember that introns and exons are present in eukaryotic genomes, but not in prokaryotes. You may wish to read p. 343 of Lehninger to review the concept of introns, exons, and protein domains. 3. What is an Expressed Sequence Tag (EST)? 4. Have a look at this DNA trace file from a human EST: http://www.ncbi.nlm.nih.gov/Traces/trace.cgi?&cmd=retrieve&val=286856102&dopt=trace&color=on&size=1&seeas=Show a) Translate this DNA to protein. You can use tools such as this one: http://web.expasy.org/translate/ Copy and paste the single letter code for the protein in 10 pt COURIER font below: b) Which protein does this DNA Expressed Sequence Tag (EST) encode? What is your evidence? c) Copy and paste the cDNA for the protein here. Use BOLD and UNDERLINE to identify the two codons on either side of the intron/exon boundary. Hint #1: there are two boundaries for you to mark because there are three exons as noted in the NCBI datafile. Hint #2: Part (d) below will be easier if you use the translate tool to prepare a copy of the cDNA nucleotides together with the amino acids. Use 10pt Courier font or smaller please. d) Open the structure of the protein in Cn3D. Highlight the two amino acids at the intron/exon boundaries. Then export the image as a PNG file and insert the image here: e) Do the locations of these two boundaries support or refute the hypothesis that exons encode protein domains? 5. Open the structure of Sperm Whale Myoglobin in Cn3D. Highlight the proximal and the distal histidines that coordinate the iron atom and bind oxygen. Export the image as a PNG file and insert it here. You may wish to resize it by dragging a corner until it fits more reasonably on the page. Using a colored pen or pencil, circle the two propionic acids on the heme group. 6. Form follows function. The three-dimensional structure of biomolecules is more conserved evolutionarily than is sequence. Why? 7. The sequences of three proteins (A, B, and C) are compared with one another, yielding the following levels of identity: A B C A 100% 65% 15% B 65% 100% 55% C 15% 55% 100% Assume that the sequence matches are distributed uniformly along each aligned sequence pair. Would you expect protein A and protein C to have similar three dimensional structures? Explain. 8. Try finding an optimal alignment for these two sequences by introducing gaps and using your knowledge of amino acid similarities to line up conservative substitutions. You may wish to do this on a computer using a text editor (Courier Font) or at least a large piece of scratch paper. MLVGSDTEKGKKIFIMECSQCHTVEKGTGPNLHGKHKGLFGRKYGYG MFTGSFTLVQAVAEATDTQRGKKVFATECSVCHAVKTGTGPNKFGYG 9. Using the NCBI website I demonstrated in class, find the sequence of the enzyme triose phosphate isomerase from E. coli. Use the FASTA format of this sequence as the query for a protein-protein BLAST search. In the output, find the alignment with the sequence of triose phosphate isomerase from Homo sapiens. How many identities are observed in the alignment? Now obtain both FASTA-formatted sequences and align them using the SLOW/ACCURATE setting of the ClustalW website at Kyoto University (http://www.genome.jp/tools/clustalw/). How many identities are observed in that alignment? 10. Myoglobin and Hemoglobin both bind oxygen. What is the function in your body of these two proteins? How do these proteins differ? And how are they similar?