SUPPLEMENTARY DATA Bioinformatic analysis of bifidobacterial
advertisement

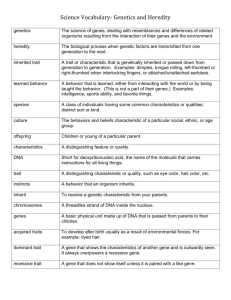
SUPPLEMENTARY DATA Bioinformatic analysis of bifidobacterial prophage genomes Prophage-like element Binf-1. The predicted prophage-like element Binf-1 in B. longum subsp. infantis ATCC15697 extends from ORF1149 (putative integrase-encoding gene) to ORF1099 (predicted holin-encoding gene) (Fig. 2). The prophage-like element Binf-1 contains a lysogeny module whose encoded proteins display significant BLAST similarities to a phage integrase (the product of ORF1149; displaying a DNA-BRE-C superfamily motif), and an anti-repressor protein (ORF1140). Notably, the lysogeny module contains two genes coding for a putative twocomponent system histidine protein kinase and a methyltransferase protein. Downstream of the lysogeny module, a small number of ORFs are present that specify proteins with similarities to proteins involved in DNA replication (e.g., recombinase proteins and primase proteins) from Rhodococcus and Mycobacterium phages. Identification of likely structural modules such as the DNA packaging genes (from ORF1116 to ORF1120) and the tail morphogenesis genes (ORF1107) was possible on the basis of observed homology to genes in high G+C Grampositive bacterial and lactococcal phages. Prophage-like element Binf-2. In this prophage-like element the lysogeny module was predicted to extend from ORF1490 to ORF1497, and includes the predicted integrase gene and genetic switch region. Downstream of the lysogeny module, a small number of anonymous ORFs and genes encoding proteins with similarities to DNA replication proteins from Enterococcus phages were identified. Database matches included a RecA ortholog, a replication initiation protein and a single-stranded DNA binding protein, defining this genome segment as the likely DNA replication module. Further downstream, several ORFs encoding hypothetical proteins were detected (Fig. 2). Their closest relatives were found in genomes from phages that infect low G+C content bacteria, such as enterobacteria, Lactococcus lactis, as well as high G+C content bacteria, such as Mycobacterium and Sinorhizobium. Notably, two adjacent DNA methyltransferase-encoding genes (ORF1526 and ORF1527) were surrounded on both sides by an antirepressor-encoding gene (ORF1525and ORF1528) and on one side by an integraseencoding gene (ORF1531), suggesting acquisition of this DNA region by horizontal gene transfer. Prophage-like element Binf-3. This prophage-like element represents what appears to be one of the most rearranged and disintegrated prophage genome identified on the genome of B. longum subsp. infantis ATCC 15697, suggesting that this prophage-like element represents a phage remnant. In fact, only modules for structural phage components and host lysis were detected (Fig. 2). The former include the putative morphogenesis genes (encoding a minor tail component, and a prohead protease), genes putatively involved in DNA packaging (encoding the large terminase subunit, as well as a portal protein), while the latter was represented by genes predicted to specify the lysin and holin (Fig. 2). The closest relatives of these DNA regions were from phages infecting Actinobacteria, such as Corynebacterium and Mycobacterium bacteriophages. Prophage-like element Binf-4. The predicted prophage Binf-4 in B. longum subsp. infantis ATCC 15697 extends from ORF1780 (integrase gene) to ORF1831 (lysin gene) (Fig. 2). Database matches allowed the subdivision of the Binf-4 prophage genome into functional modules (Fig. 2). In this prophage-like element, the lysogeny module is limited to a region extending from ORF1780 to ORF1784, and encompassing genes that specify homologs of the integrase of Leifsonia xyli and the cI repressor of M. tuberculosis. The next ORFs (ORF17911795) constitute the likely DNA replication module, as it includes genes whose encoded protein products display similarity to single-stranded DNA binding proteins of mycobacteriophage, the partitioning protein of Clostridium, a putative replication protein of an Enterobacterium phage and an oligoribonuclease. Identification of structural modules such as the DNA packaging genes and head morphogenesis genes (from ORF1809 to ORF1815), and tail morphogenesis genes (from ORF1821 to ORF1823) was possible on the basis of observed homology to genes present in genomes of high G+C mycobacterial prophages. The general organization of the regions spanning part of the tail fiber and the host lysis cassette (from ORF1831 to ORF1832) showed a one to one correspondence to similarly sized genes in the tail fiber and host lysis clusters of the Binf-3 prophage-like element (Fig. 2). Notably, the lysis cassette, which is constituted by the lysin and holin-encoding genes, was identified downstream of the tail fiber module. The order of the lysin and holin-encoding genes is opposite to what is generally described for prophage genomes (Canchaya et al., 2003) and this genetic constellation is conserved in all identified B. longum subsp. infantis prophage genomes. Prophage-like element Binf-rmn-3. As shown in Figure 2, the genetic structure of Binf-rmn-3 appears to have been the result of multiple DNA deletion and rearrangement events. In fact, its small size and the absence of a region encoding structural phage components suggest that one or more major DNA deletion events have occurred in this prophage-like element. Prophage-like element Binf-5. The 37,747 bp genome of this prophage-like element extends from ORF2467 to ORF2406, which encode an integrase protein and a phage holin, respectively. This genome was predicted to contain functional modules such as the lysogeny module (ORF2467 to ORF2459) with a typical genetic switch region, and DNA replication module (ORF2456 to ORF2447) (Fig. 2). Putative tail morphogenesis and head morphogenesis genes were identified on the basis of observed homology to similar genes in high G+C Corynebacterium phages. The rightmost part of the Binf-5 genome represents a likely host-lysis module, comprised of the holin (ORF2406) and lysin (ORF2407) genes showing database matches to homologous genes of the B. longum subsp. longum Blj-1 prophage (Ventura et al., 2005). Prophage-like element Binf-6. In this prophage-like element the lysogeny module is limited to a phage integrase gene (ORF1137), and to a superinfection exclusion gene (ORF1133) whose deduced protein product shares significant similarity with the Lp1 prophage superinfection immunity gene of Lactobacillus plantarum (Ventura et al., 2003). The lysogeny module is followed by a DNA replication module containing genes that are predicted to encode an oligoribonuclease (ORF1119) and a primase (ORF1098). The next ORFs (ORF1090 to ORF1078) constitute the likely morphogenesis module. Notably, the Binf-6 prophage-like element contains a transposon-like element encompassing a transposase (ORF1075) and a putative site-specific integrase/resolvase (ORF1073) resembling a B. longum subsp. longum DJO10A transposon. The presence of such a composite mobile element inserted into a prophage genomic sequence is not unusual. In fact, in B. breve UCC2003 a prophage genomic sequence has been described that carries a transposon element encompassing genes encoding a putative ABC transporter system and two glycosyl hydrolases (Ventura et al., 2005). The transposon element in the Binf-6 prophage is followed by ORF1070-1069 and ORF1067, encoding hypothetical products and a minor tail protein, respectively. Prophage-like element Binf-7. The analysis of the genome of B. longum subsp. infantis CCUG52486 revealed, apart from Binf-6, one additional prophage-like element, Binf-7. The genomic organization of this prophage-like element resembles that of the B. longum subsp. longum Bl-1 prophage (Ventura et al., 2005). The Binf-7 integrase gene is preceded by a predicted holin and a lysin gene with sequence similarity to the B. longum subsp. longum prophage Bl-1. Of note, the deduced lysin sequence contains a typical amidase motif (pfam01510). Further upstream, putative morphogenesis genes (encoding a minor tail component, and a prohead protease) and genes putatively involved in DNA packaging (encoding a terminase, as well as a portal protein) were identified (Fig. 2). Prophage-like element Bbif-1. The genome organization of Bbif-1 resembles a typical lambdoid phage genome structure. Downstream of the lysogeny module, several ORFs are predicted to encode proteins with homology to DNA replication proteins, thus defining this genome segment as the likely DNA replication module. Further database matches allowed a distinction of likely DNA packaging and head morphogenesis genes, possible head-to-tail joining genes and putative tail genes. A lysis cassette was identified downstream of the tail fiber module, which is constituted by the classical genetic constellation of a holin and lysin gene. Prophage-like element Bbif-2. Database matches allowed the identification in the genome of Bbif-2 prophage-like element of ORFs displaying similarities to phage-related genes, some of which could be clustered into functional modules (Fig. 3). In this prophage-like element, the lysogeny module is limited to a region spanning ORF1254 to ORF1273. The rest of the Bbif-2 genome contains ORFs displaying similarities to morphogenesis genes but apparently distributed in a non-modular fashion. Only the rightmost part of the genome contains a typical host lysis module which encompasses the presumptive holin gene (ORF1338) and an endolysin gene (ORF 1339). Prophage-like element Bado-1. The predicted integrase-encoding gene (ORF958) is located next to the host lysis cassette in a genetic constellation that is similar to that of Blj-1 in B. longum subsp. longum DJO10A strain (Ventura et al., 2005). The database matches of the predicted ORFs allowed a distinction of DNA packaging and head morphogenesis genes producing the highest similarity scores with the analogous genes (DNA packaging and head morphogenesis) from phages infecting Mycobacterium and Streptomyces. Notably, a small number of genes that encode proteins with similarity to bacterial proteins were identified between the host lysis cassette and the predicted morphogenesis module. Notably, these include a complete Type I R/M system and genes involved in polyketide biosynthesis (e.g., betalactamase and enoyl-CoA hydratase) (Fig. 5). Prophage-like element Bado-2. The predicted prophage Bado-2 extends from ORF1182 (integrase gene) to ORF1212 (putative terminase) on the genome of B. adolescentis L2-32. The genome sequences of this prophage includes ORFs encoding predicted structural components of the phage and genes of the lysogeny module (encoding a predicted integrase and Cro-like protein), host lysis cassette (lysin gene) and two transcriptional regulators (ORF1189 and ORF1193) (Fig. 5). Prophage-like element Bado-3. The genome architecture of prophage Bado-3 resembles that of Bado-2, despite the lack of significant DNA similarities between these two prophage genomes (Fig. 5). The lysogeny module of Bado-2 includes two genes (ORF1370 and ORF1372) whose deduced protein products show a high level of identity to an integrase and a cI-like repressor, respectively. Two adjacent genes (ORF1373 and ORF1374) were assigned to the host lysis cassette, which is constituted by the classical genetic constellation of holin and lysin-encoding genes. Further database matches allowed the distinction of likely DNA packaging and head morphogenesis genes, putative head-to-tail joining and tail genes. Prophage-like element Bdent-1. The predicted prophage-like element Bdent-1 extends from ORF1446 (integrase gene) to ORF1477 (lysin gene) (Fig. 6). In this prophage-like element, the integrase-encoding gene is placed immediately upstream of the likely DNA replication module consisting of two ORFs (ORF1450 and ORF1453), which encode proteins with similarities to a primase and DNA helicase from Firmicutes. Identification of likely structural modules such as the head morphogenesis and DNA packaging (from ORF1460 to ORF1462) and tail morphogenesis genes (from ORF1469 to ORF1472) was possible on the basis of observed homology to lactococcal and streptococcal phages. Further upstream, a gene (ORF1474) similar to an endopeptidase detected in the genome of Streptococcus pyogenes was identified, which could be part of a structural tail component required for cell wall lysis to allow phage entry during infection (Kenny et al., 2004); separated from this gene by an anonymous ORF, a gene encoding a putative lysin (ORF 1477) was identified. Prophage-like element Bdent-2. Database matches indicated that the Bdent-2 prophage genome is subdivided into functional modules (Fig. 6). In this prophage-like element the lysogenic module is limited to a phage integrase gene (ORF0601), whose deduced protein product displays significant similarity to phage integrases of Mycobacterium and Corynebacterium species. The integrase-encoding gene is followed by genes putatively involved in DNA packaging (encoding a small and large terminase subunit, as well as a portal protein), and putative tail morphogenesis genes (encoding a minor tail component, and a prohead protease). Their closest relatives are from phages infecting low G+C content bacteria like L. lactis bIL170, Lactobacillus delbrueckii bacteriophage LL-H and from phages infecting high G+C content bacteria like Mycobacterium and Corynebacterium. Further upstream, two genes (ORF1598 and ORF1604) are located that encode proteins with weak similarity to an excisionase protein of Frankia and a cI-like repressor of Listeria (Fig. 6), thus constituting the predicted lysogeny module. References Canchaya, C., Proux, C., Fournous, G., Bruttin, A., and Brussow, H. (2003). Prophage genomics. Microbiol Mol Biol Rev 67(2), 238-76, table of contents. Kenny, J. G., McGrath, S., Fitzgerald, G. F., and van Sinderen, D. (2004). Bacteriophage Tuc2009 encodes a tail-associated cell wall-degrading activity. J Bacteriol 186(11), 3480-91. Ventura, M., Canchaya, C., Kleerebezem, M., de Vos, W. M., Siezen, R. J., and Brussow, H. (2003). The prophage sequences of Lactobacillus plantarum strain WCFS1. Virology 316(2), 245-55. Ventura, M., Lee, J. H., Canchaya, C., Zink, R., Leahy, S., Moreno-Munoz, J. A., O'Connell-Motherway, M., Higgins, D., Fitzgerald, G. F., O'Sullivan, D. J., and van Sinderen, D. (2005). Prophage-like elements in bifidobacteria: insights from genomics, transcription, integration, distribution, and phylogenetic analysis. Appl Environ Microbiol 71(12), 8692-705.