Model Manuscript (IJBSANS)
advertisement

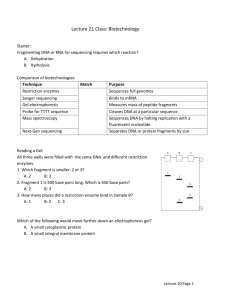
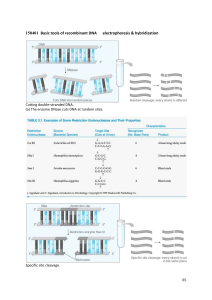
“Model Manuscript (IJBSANS)” Partial purification and Insilico analysis of Bst1 from Bacillus stearothermophilus xxxxxxxa*, xxxxxxxxb*, xxxxxxc, xxxxxxd, xxxxxe, and xxxxxxxxxd#. a xxxxxxx china xxxxxxx India. c xxxxxxxxxxxxxxxxxxxx USA. d xxxxxxxxxxx Italy. e xxxxxxxx Germany b * These authors are co-first authors # Corresponding author xxxxxxxx xxxxxxxx Email: xxxxxxxxxx 1 Abstract: There are more than 3000 restriction endonucleases which cut the DNA in their respective sites. In the present study, the thermostable restriction enzyme BstI was partially purified from Bacillus stearothermophilus. The phosphocellulose column purified Bst1 elutes was standardized to digest E.coli, plasmid and λ DNA. Further, an in silico analysis was done using NETCUTTER, a bioinformatics tool to generate restriction map to match the fragment sizes we obtained in the agarose gel electrophoresis. Keywords: Bacillus stearothermophilus, BstI, phosphocellulose, DNA, insilico Introduction The discovery of the enzymes which led to Nobel Prize for Artbar and Nathans in 1978 was one of the key breaks through in the development of genetic engineering. By definition, restriction endonucleases are part of restriction modification systems (RM), which comprise an endoculease and methyl transferase activity (Pingound and Jeltsch, 2001). Type of RM system have been found and were classified according to their subunit composition, co-factor requirements and mode of action (Simth and Nathans, 1973) the distinction between type I,II and III system is still useful that there are intermediate cases. Most characterized enzymes fall into type II class that is further divided into many subclasses have been characterized primarily with respect to their recognition sequence and cleavage specificity rather than their proteins properties. All restriction endonuclases cleave their DNA substrate to form 5’Phosphate and 3’Hydroxyl termini on each strand. The first thermostable restriction endouclases was reported was TaqI from the North American T. aquaticus type stains YT (Sato et al., 1977). All structure of restriction endonuclease show a common structural are comprising four β-stands and one αhelix (Pingound and Jeltsh, 2001). Site specific endonuclease is currently indispensable tools in the studies of DNA structure and other molecular genetic studies. There are about more than 3000 type II restriction enzymes available at the market now. The range of available enzymes should be expanded to provide further progress in this research field. The quality of enzymes is an important as the range of their choice. Therefore the development of effective technologies of preparation of restriction endonuclease and also to prepare thermophilic restriction enzymes was urgent problem. The goal of the present study was to develop a small scale production and purification of BstI restriction endonuclease from Bacillus stearothermophilus. 2. Materials and Methods: 2.1 Strains Bacillus stearothermophilus (MTCC 1521) strain was purchased from MTCC, Chandigarh, India. 2.2 Production media for Bacillus Sterostermophilius Bacillus sterostermophilius were grown in a modified SLBH medium contains [for 100ml: 1.1g- tryptone, 2.25g - yeast extract, 5.1ml- (1M) K2PHO4, 1.57(1M) - KH2PO4, 0.4mlglycerol] this medium kept in 60°C h in a shaker. 2 2.3 Production of restriction enzymes Culture grown till late log phase was centrifuged at 6000rpm 15 min at 4°C. The supernatant was discarded and the pellet was collected and stored at -20°C for 2 hours. Forzen cell pellets were thawed and suspended in 10ml of buffer A (10mM potassium sulfonylfloride pH 7.0, 1mM EDTA, 1mM 2-mercaptoethanol) containing 2.5mg phenylmethyl sulfonylfluoride (PMSF) and 0.4M NaCl. Sometimes it can be effective without proteolysis inhibitor (PMSF). However this was generally added as a precaution. The cell suspension was sonicated with the cell suspension on ice was carried out in 30’-90’ pulse; the temperature was monitored and kept below10°C strains which were resistant to sonication. These were stirred on ice with the addition of 10µl/ml lysozyme for 15-30 min before sonication. They were sonicated for cell repture for 50cycle of 2 min each with a time gap of 5 min between each cycle. The sonicated cell suspension was centrifuged for 1 hour at 12,000 rpm in 4°C. The supernatant was collected as crude of restriction enzyme. 2.4 Purification of restriction endonucleases 2.4.1 Ammonium Sulphate Preparation The crude enzymes were precipitated using the standard salting out procedure using solid ammonium sulphate using 80% saturation and stirred well for 2h at 4°C and centrifuged at 20,000rmp for 1h. The supernatant was removed and the precipitated formed was dissolved in buffer A contains 0.1M NaCl. 2.4.2 Dialysis The sample from ammonium sulphate precipitation was transferred to in dialysis membrane bag with 1000MV cut off dialysis tubing and dialyzed against buffer A containing 0.1M NaCl at 4°C for 18h. The buffer was changed every 4 h. 2.4.3 Phosphocellulose chromatography Careful preparation of phosphocellulose is essential to the success of this purification method. 1g of phosphocellulose were suspended in 32ml of 0.2N HCl diluted 1:1 with 95% ethanol and stirred gently for 30min at room temperature and filtered solution using Whatemann filter paper. The slurry was allowed to settle and the supernatant aspirated or decanted to remove fine and other particulate matter. The collected resin was washed 2 to 3 times by suspension in 20ml of distilled water. The pH was adjusted to near neutral with 1M NaOH and the resin was suspended in 50ml 0.1N NaOH stirred for 30min at room temperature, collected by filtration and suspended in 50ml of 1mM EDTA. It was stirred for 30min at room temperature and washed 3 times with distilled water. The pH was adjusted to near neutral with 1M HCl and the resin was suspended in extract buffer plus 0.2M NaCl. The pH was carefully adjusted to pH 7.0 before pouring a column since many column volume of dilute buffer and required for equilibration of the pH in a packed column. The phosphocellulose column (1.2X15cm) is sufficient for cell. It should be washed with several volumes of extract buffer-A prior to loading. The extract was loaded directly on to the column and the column washed with several column volumes of extract. The restriction enzyme was eluted with the buffer-A containing 0.4M NaCl. Ten fractions of 1ml were collected and assayed for endonucleae activity and protein estimation. 2.5 Genomic and plasmid DNA isolation from bacteria A single colony of Escherichia coli DH5 α cell containing pGEM 7Z f+ was inoculated in to 3ml of LB medium containing ampicillin (100µg/ml) and was grown with constant shaking at 37°C for 16h. The plasmid DNA was isolated according to standard protocol Sambrook, et al., (1989). 3 2.6 Restriction Assay with λ and E.coli Genomic DNA Two vials were used for E.coil and λ DNA Restriction assay. The reaction was set in 30µl of reaction mixture containing 3µl of assay buffer (100mM Tric HCl pH 7.0, 10mM MgCl2, 10mM NaCl), 1µl of DNA (E.coli DNA in one tube and λ DNA in another tube) and 21µl of crude, dialyzed and purified restriction enzyme BstI. The reaction mixture was incubated 24h at 37°C and the reaction was stopped after incubation time. The fragments were analyzed using 1.0% agarose gel electrophoresis containing ethidium bromide (2µl/ml). The bands were visualized under UV illuminator and were documented using a gel documentation system. 2.7 Restriction assays for plasmid DNA Plasmids isolated from Staphylococcus aureus were digested using BstI and 24 h at 37°C. The fragments were analyzed using 1% agarose gel electrophoresis. 2.8 In Silico analysis Using bioinformatics tool, NET CUTTER, restriction map was generated for Bst1 with λ and E.coli DNA to compare with size of bands of BstI observed in agarose gel electrophoresis. 3. Results 3.1 Production of Restriction enzymes Bacillus stearothermophilus grown in SLBH medium at 65°C for 6h showed maximum activity for BstI. Similarly, Salleh, et al. (1977) isolated proteases from Bacillus stearothermophilus which is stable at 60°C and Razak, et al., (1993) reported that Bacillus sp. has produced a thermostable protease that has an optimum activity at 60°C. 3.2 Purification of BstI Phosphocellulose column was used for the purification of BstI. The purification profile of BstI showed 72.72% yield and 514 fold increase in purification. Highest specific activity was obtained in third fraction of phosphocellulose column and the specific activity of 10 fractions was shown in Figure 1. Catterall and Welker, (1977) investigated the small scale purification of class II-endonuclease (BstI) from strain NCA 1503. 3.4 Restriction analysis of lambda, E.coli, and plasmid DNA Restriction assay of BstI with λ and E.coli DNA showed 22 sites and 20 sites respectively (Fig 2). This result was cross checked using bioinformatics analysis (Table 1). In order to confirm the cutting efficacy of purified BstI, we analyzed this enzyme to restrict plasmids isolated independently by one of our colleague. This restriction analysis showed BstI indeed cut in its recognition sites as observed by many bands in the agarose gel electrophoresis (Fig.2). 4. Discussion Similarly, Salleh, et al. (1977) isolated proteases from Bacillus stearothermophilus which is stable at 60°C and Razak, et al., (1993) reported that Bacillus sp. has produced a thermostable protease that has an optimum activity at 60°C. Catterall and Welker, (1977) investigated the small scale purification of class II-endonuclease (BstI) from strain NCA 1503. This result was cross checked using bioinformatics analysis In order to confirm the cutting efficacy of purified BstI, we analyzed this enzyme to restrict plasmids isolated independently by one of our colleague. This restriction analysis showed BstI indeed cut in its recognition sites as observed by many bands in the agarose gel electrophoresis. 4 5. Conclusion In conclusion, restriction enzymes are indispensable tools in the molecular biology studies the standardization of the procedure for isolating and obtaining high purify enzymes are very important. Bacillus stearothermophilus (MTCC 1521) was tested for the production of restriction enzymes. Ammonium sulphate precipitation and dialysis were used for the partial purification of the restriction enzymes preparations. Phosphocellulose column which is not only an anion exchange but also an affinity column for DNA binding proteins was used for the final level of purification. The BstI was partially purified and fractions were standardized to assay with E.coli, λ DNA and plasmid DNA and restriction sites were confirmed using insilico analysis. BstI can be used as an alternative for Sau3AI and MboI because of its same recognition sequence. 5. REFERENCE: 1. Alfered Pingound and Albert Jeltsch, (2001). Structure and function of Type II Restriction enzymes. Nucleic Acid Research, 29, 18, 3705-3727. 2. Simth, H.O. and Nathans, D. (1973). A suggested ammenclature for bacterar hast medification and restriction system and other enzymes. Journal of Molecular Biology, 18, 419-423. 3. Sato S and Hutchison CA, et al., (1977). A thermostable sequence-specific endonuclease from Thermus aquaticus. Proc Natl Acad Sci. 74:542–546. 4. Sambrook,J and Fritsch EF, et al., (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 5. Salleh AB and Basri , et al., (1977). The effect of temperature on the protease from Bacillus stearothermophilus strain F1. Mal. J. Biochem. Mol. Biol. 2, 37-41. 6. Razak, C and Samad, M, et al., (1993). Thermostable extracellular protease by B. stearothermophilus. World J. Microbiol. Biotechnol. 10, 260–263. 7. Catterall, JF and Welker NE, (1977) Isolation and properties of a thermostable restriction endonuclease (ENDO R-Bst1503)J. Bacteriol. 129, 1110-1120. 5 Figure1: Specificity Activity of different fraction of BstI 500 A c tiv it y 400 300 200 100 0 1 2 3 4 5 6 F r a c t io n s 6 7 8 Figure 2: Restriction of BstI with E. coli DNA (a) lambda DNA (b) Line 1: Marker DNA Lane 2: BstI with lambda DNA 7 Table 1: Analysis of restriction site S.NO Site Bst Target 1 AGCTC 2 2 ACGTCAG 3 8