Summary
advertisement

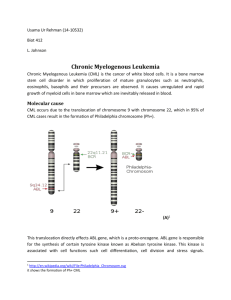
Summary Chronic myeloid leukemia (CML) is amyeloprolirative disorder characterized by the Philadelphia chromosome originates from the translocation of chromosome 9 and 22 t(9;22) (q34;q11), creating a BCRABL fusion gene which results in the constitutive activation BCR-ABL tyrosine kinase that triggers leukemogenic pathways resulting in onset of chronic myeloid leukemia. There are different forms of the BCR-ABL fusion gene including b3a2, b2a2 codes for p210 protein and e1a2 fusion gene codes for p190 protein. Imatinib mesylate is the effective therapy that inhibits the tyrosine kinase activity of BCR-ABL as a first line treatment for all newly diagnosed chronic phase CML patients. By binding to the ATP binding pocket, it prevents ATP from binding site and the phosphorylation of downstream substrate is disturbed. However, drug resistant have been founded and the most important reasons for this resistant are point mutations in the kinase domain. These mutations alter the conformation of the ATP binding site, disturb the binding of therapy and lead to Imatinib resistance. The present study deals with different aspects: conventional cytogenetic (G-banding) analysis to diagnose the Philadelphia chromosome as a CML marker, more sensitive molecular techniques (RT-PCR) were conducted as a confirmation tests after cytogenetic assay to detect different types of BCR-ABL fusion genes also to monitor of molecular remission finally using Allele Specific Oligonucleotide Polymerase Chain Reaction (ASO-PCR) assay for screening the mutant allele at the three codons 351, 311 and 315of the BCR-ABL ATP binding domain in Imatinib resistant CML patients. In the present study phenotype of fifteen CML patients were successfully analyzed, thirteen patients were in complete cytogenetic response state (Philadelphia 0%) only two of those patients show the philadelphia chromosome then the negative samples were subjected to multiplex reverse transcriptase polymerase chain reaction to analysis the variant translocations. Out of fifteen patients examined nine CML cases were positive for BCR-ABL transcripts. Two patients express b3a2, three patients express b2a2 while one patient express e1a2 and one express b2a3. On other hand we detect co-expression of transcripts like two CML cases showed e1a2 and b2a3 co-expression and one patient showed e1a2 and b3a3 co-expression. Among the nine positive fusion genes eight of them were showed positive BCR-ABL transcripts by in Nested reverse transcriptase polymerase chain reaction that express b3a2, b2a2 and e1a2 fusion genes. Three point mutations T1052C, T932C and C944T were identified by allele specific oligonucleotide polymerase chain reaction. One patient showed T1052C mutation, T932C was detected in three patients and two patients showed the C944T ATP binding domain mutation. In conclusion, conventional cytogenetic and molecular genetic tests are complementary techniques to show the exact types of abnormalities, that allow better evaluation of the genomic aberration involved in CML patients, specifically for kinase domain mutation that plays an important role in different diagnosis, prognosis, therapy treatment and drug resistant management of chronic myeloid leukemia.