X-ray crystallography - How it works and what it is used for.
James Watson 1964: “Unfortunately, we cannot accurately describe at the chemical level how a molecule
functions unless we know first its structure”
First we will watch a cartoon.
http://richannel.org/a-case-of-crystal-clarity
Now we will see how to grow and examine a protein crystal to get the protein structure.
http://richannel.org/understanding-crystallography-part-one
3D structure of a protein called lysozyme.
It's an enzyme that's in our tears and saliva and mucus, and it helps us fight bacteria.
3D structure helps us to understand the mechanism of action of this enzyme, and therefore how it helps
fight bacteria.
The wavelength of light is much, much larger than the size of this tiny molecule.
So we use X-rays to look at these molecules.
In each protein crystal are millions of protein molecules, all arranged in an ordered, grid-like structure.
By firing X-rays at these, and measuring how they scatter, we can work out the molecular structure of
nearly any crystallized sample.
X-ray crystallography - some of the most important biological structures have been obtained: the double
helix of DNA to numerous proteins, vitamins, and drugs.
It can take a long time to grow a suitable crystal.
Genetically modify E. coli bacteria, which then produces protein.
Break open the cells, extract the protein, and then purify it.
Grow crystals of the protein molecules.
Get the protein molecules to line up in an ordered array-- such as in a crystal, where they're all lined up in
the same orientation-- when the X-rays scatter from the crystal, then we can get information.
The signal is strong enough to get the three dimensional structure of the protein.
The protein structures we work on today are far more complex.
They can produce very small and delicate crystals.
They need extremely powerful X-ray sources at specialist facilities.
- See more at: http://richannel.org/understanding-crystallography-part-one#bm_23250
28
http://richannel.org/understanding-crystallography-part-two#bm_11170
A particle accelerator that can generate beams of light 10 billion times brighter than the sun.
Users are structural biologists doing x-ray crystallography.
In the crystal, the ordering of the molecules means that when we fire x-rays at a crystal, it diffracts or
scatters the beam into hundreds of intense rays.
The resulting diffraction pattern, detected as an array of spots, is dependent on the internal structure of
the crystal.
By measuring the intensities and positions of these spots, we can work back to the structure of the
molecule it is made from.
A pulse of x-rays shoots straight into one of the lead-lined experimental hatches where scientists carry out
their diffraction experiments.
During the experiment, the crystal is cooled by ultra-cold nitrogen gas to help it withstand the radiation.
The diffracted rays are captured on a detector.
X-rays are a form of light that has a wavelength about the size of an atom.
Mathematical understanding of how light scatters allows scientists to work backwards from the diffraction
pattern to a three-dimensional image of the protein in the crystal.
This gives three-dimensional maps, which plot the electron density of the protein molecule.
The resolution of these maps is dependent on the quality of the crystal.
With good quality data, these maps are very detailed. You can see the lumps and bumps of every atom,
every twist and turn of the protein chain.
From these sorts of investigations, we can learn how the biological molecules of your body interact with
one another, or how to design drugs that will bind to them more effectively, or even to design proteins
that nature never got around to inventing.
Korea has a synchrotron at Pohang.
The Pohang Light Source was designed to provide synchrotron radiation with continuous wavelengths
down to 1Å. It's construction was completed in September, 1994. The PLS is a national user facility, owned
and operated by the Pohang Accelerator Laboratory (PAL) and POSTECH on behalf of the Korean
Government.
Now we can study how this information might be used.
29
Structure of Ribosomes
Structure determination by X-ray crystallography
An X-ray structure of a bacterium ribosome. The rRNA-molecules are colored orange, the proteins of the
small subunit are blue and the proteins of the large subunit are green. An antibiotic molecule (red) is
bound to the small subunit. Scientists study these structures in order to design new and more effective
antibiotics.
Different antibiotics can be used in the fight against disease-generating bacteria. Many of these antibiotics
kill bacteria by blocking the functions of their ribosomes. However, bacteria have become resistant to most
of these drugs more often. Therefore we need new ones.
X-ray crystallography structures show how different antibiotics bind to the ribosome. Some of them block
the tunnel through which the growing proteins leave the ribosome, others prevent the formation of the
peptide bond between amino acids. Still others stop the correct translation from DNA/RNA-language into
protein language.
Several companies now use the structures of the ribosome in order to develop new antibiotics
http://www.nobelprize.org/nobel_prizes/chemistry/laureates/2009/popular-chemistryprize2009.pdf
30
The development of Imatinib
The classic example of targeted development is imatinib mesylate (Gleevec), a
small molecule which inhibits a signaling molecule kinase.
The genetic abnormality causing chronic myelogenous leukemia(CML) has
been known for a long time to be a chromosomal translocation creating an
abnormal fusion protein, kinase BCR-ABL, which signals badly, leading to
uncontrolled proliferation of the leukemia cells.
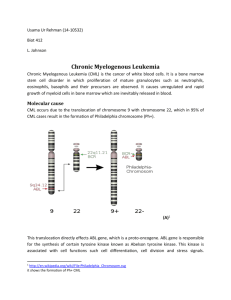
The Philadelphia chromosome
Discovered in 1960, the diagnostic karyotypic abnormality for chronic myelogenous leukemia is
shown in this picture of the banded chromosomes 9 and 22. Shown is the result of the reciprocal
translocation of 22q to the lower arm of 9 and 9q (c-abl to a specific breakpoint cluster region [bcr] of chromosome 22 indicated by the arrows). ©
2006 Peter C. Nowell Courtesy of Peter C. Nowell, MD, Dept. of Pathology and Clinical Laboratory of the University of Pennsylvania School of
Medicine. All rights reserved.
In 1983, researchers demonstrated that the human c-abl oncogene is located in the region of chromosome 9 that
translocates to become part of the Philadelphia chromosome.
in 1990, scientists infected bone marrow cells with a retrovirus encoding the fusion gene and demonstrated that,
indeed, the presence of an active bcr-abl gene initiates CML or CML-like symptoms
in 1990, researchers identified the function of the bcr-abl fusion gene: production of an abnormal tyrosine
kinase protein that is not properly regulated. Tyrosine kinase is a common signaling molecule that, when activated,
triggers cells to divide. In patients with CML, the mutated tyrosine kinase is active for far too long, causing cells to
proliferate at an abnormally high rate. This proliferation results in the overproduction and accumulation of immature
white blood cells, which is the sign of CML. A milliliter of blood from a healthy person contains about 4,000 to 10,000
white blood cells, the same volume of blood from a person with CML contains 10 to 25 times that amount.
Scientists learned about kinase structure, and found considerable variation among the different kinases with respect
to the structure of their ATP-binding pockets. This meant that a drug that only blocks the bcr-abl ATP binding site
might be possible.
Ciba-Geigy (which later became Novartis), were conducting tyrosine kinase inhibitor research. They had already
synthesized some kinase-blocking inhibitor compounds, using computer models to predict which molecular
structures might fit the ATP-binding site of the fusion protein.
Imatinib precisely inhibits this kinase.
Brian Druker had researched the abnormal enzyme kinase in CML.
He thought that precisely inhibiting this kinase with a drug would
control the disease and have little effect on normal cells.
Druker collaborated with Novartis chemist Nicholas Lydon, who
developed several candidate inhibitors.
From these, imatinib was found to have the most promise in laboratory
experiments.
Druker found that when this small molecule is used to treat patients
with chronic-phase CML, 90% achieve complete haematological
remission.
It is hoped that molecular targeting of similar defects in other cancers
will have the same effect.
31
Imatinib is marketed by Novartis as Gleevec or Glivec. It is a tyrosine-kinase inhibitor used in the treatment of
multiple cancers.
Like all tyrosine-kinase inhibitors, imatinib works by
preventing a tyrosine kinase enzyme, in this case
BCR-Abl, from phosphorylating subsequent proteins
and initiating the signalling cascade necessary for
cancer development, thus preventing the growth of
cancer cells and leading to their death by
apoptosis. Because the BCR-Abl tyrosine kinase
enzyme exists only in cancer cells and not in healthy
cells, imatinib works as a form of targeted therapy—
only cancer cells are killed through the drug's
action. Imatinib was one of the first cancer therapies
to show the potential for such targeted action, and is
often cited as a paradigm (very good example) for
research in cancer therapeutics.
Crystallographic structure of tyrosine-protein kinase ABL (rainbow colored, Nterminus = blue, C-terminus = red) complexed with imatinib (spheres, carbon =
white, oxygen = red, nitrogen = blue).
Here is an animation of how Gleevec works.
http://www.dnalc.org/resources/3d/32-how-gleevec-works.html
Rational drug design
Flow chart for structure based drug design.
By Laozhengzz CC A-SA 3.0 http://en.wikipedia.org/wiki/File:Flow_chart_for_structure_based_drug_design.jpg
References:
Pray, L. (2008) Gleevec: the breakthrough in cancer treatment. Nature Education 1(1):37
http://www.nature.com/scitable/topicpage/gleevec-the-breakthrough-in-cancer-treatment-565#
Imatinib http://en.wikipedia.org/wiki/Gleevec
32