mec5629_sm_AppendixS3
advertisement

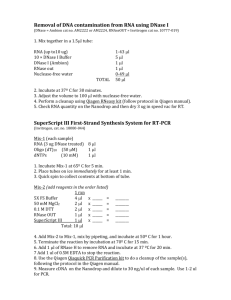
Medicago-Sinorhizobium GeneChip procedures After the supplementary protocol of Barnett et al. 2004. PNAS 101: 1663616641 and the Affymetrix expression_analysis_technical_manual.pdf, but using different RNA isolation and DNA removal protocols. Reagents and items needed: Forceps for removing nodules Liquid nitrogen (LN2) Dry ice Mortar & pestle TRI REAGENT (Molecular Research Center, Inc #TR 118) Sturdy polypropylene microcentrifuge tubes RNase-away (Invitrogen, #10328011) RNase-free water RNase-free water, pH adjusted to 7.0 with NaOH 100 bp DNA ladder (New England Biolabs #N3231S) Turbo DNA-free kit (Ambion #AM1907) DNAse I (Pierce #27-0514-0 or Fisher #PI-89835) One-phor-all 10X buffer (USB #78331) Qiaquick PCR Purification Kit + buffer PB for cDNA cleanup (Qiagen #28104) Random primers (Hexamers, 75 ng/µl, Invitrogen #48190011) Superase Inhibitor (Ambion #AM2694) Superscript III (200 unit/µl Invitrogen #18080044) with 100mM DTT buffer and 5x first-strand buffer: 250 mM Tris pH 8.3, 375 mM KCl, 15 mM MgCl2 10 mM dNTPs (Invitrogen #N80800078) I N NaOH I N HCl GeneChip DNA Labeling Reagent (Affymetrix #900542) Terminal Deoxynucleotidyl Transferase, Recombinant (Promega #M1871) Terminal Transferase, Recombinant, is supplied in 50mM potassium phosphate (pH 6.4), 100mM NaCl, 1mM β-mercaptoethanol, 0.1% Tween® 20 and 50% glycerol Terminal Transferase 5X Buffer: 500mM cacodylate buffer (pH 6.8), 5mM CoCl2 and 0.5mM DTT Poly-A RNA controls (Affymetrix #900433) All supplies and reagents must be RNAse-free at all steps until cDNA is made. Gels for DNA may be 1.5% agarose in 0.5x TBE buffer. The tracking dyes will have apparent molecular weights of ~4kb (xylene cyanol FF), ~ 300bp (bromophenol blue), ~50bp (Orange G). Choose dyes that do not obscure your sample. RNA may be run on the same gels, although bands will not be a sharp as for RNA run on formaldehyde gels. 1 Isolation of Nodule RNA Grow plants as desired. At least 800 mg of nodules are needed for a single Affymetrix GeneChip. Before harvest, label 50-ml centrifuge tubes and pack in dry ice. To excise nodule material, pinch the nodule with forceps at the nodule-root junction. Periodically add more liquid nitrogen to the tube while harvesting. Keep tubes on dry ice until nodule harvest is completed, then cap and transfer to liquid nitrogen for transport. Tissue may be stored at -80°C or used immediately. See Table 1 below for the plant samples used in this experiment. A. Total RNA Extraction: 1. Freeze mortar and pestle completely before adding nodules (pack mortar in dry ice; after mortar and pestle are cold add LN2) 2. Add nodule tissue from both samples to be pooled (2 plants worth of nodules), and grind to fine powder (repeatedly adding LN2 to keep frozen) 3. Add 3mL TRI Reagent directly to frozen mortar/pestle and tissue, and grind to a fine powder; remove mortar from dry ice bed and continue grinding as material thaws. This is period of maximum RNA extraction. 4. Split homogenized tissue into 3 tubes, each with ~1.0 mL TRI Reagent mix and proceed per tube: 5. Centrifuge TRI Reagent preps at 12,000 g for 10 minutes at 2-8 C to precipitate tissues. RNA is in supernatant. Transfer cleared homogenate to a fresh tube. 6. Incubate tubes at room temp for 5 minutes. 7. Phase separation: add 0.2 mL chloroform to each tube (0.2 mL per 1 mL TRI Reagent) & shake vigorously for 15 seconds. Incubate for 2-3 minutes at room temp. 8. Centrifuge at 12,000 g for 15 minutes at 2-8° C for phase separation (RNA is in upper, aqueous phase). Transfer aqueous phase to fresh tube (~ 900 L aqueous recovered). 9. RNA precipitation: Add 0.50 mL isopropanol to aqueous phase (0.5 mL per 1 mL TRI Reagent). 10. Incubate samples at 15-30° C for 10 minutes, then centrifuge at 12,000 g for 10 minutes at 2-8° C. RNA precipitate forms gel-like pellet on bottom/sides of tube. 11. Remove supernatant and wash pellet with cold 1 mL 75% ethanol. Vortex and centrifuge at 7,500 g for 5 minutes at 2-8° C. Decant ethanol and invert tube on clean kimwipe (removing as much EtOH as possible – carefully tap tubes on kimwipe repeatedly). 12. Briefly dry the RNA pellet by air-drying at room temp for 3-5 minutes in the laminar flow hood. (don’t overdry, as this will reduce RNA solubility). Sniff to make sure EtOH is gone. 13.Resuspend in 50 μl RNase-free water, incubate 10 min. at 55-60° C and store RNA at -80° C to minimize degradation. 2 Quantify RNA spectrophotometrically. Ideally, 260/280 ratio should be >1.8. Run RNA gel. Hope to see 4 clear bands for rRNAs, little smearing and little degradation (low molecular weight smear). See Figure 1 for example agarose gel. B. Turbo DNA-free kit for genomic DNA contamination of RNA samples. (product Ambion AM1907, protocol modified from manufacturer’s) Transfer RNAs to 0.5 mL tubes, to facilitate removal of supernatant. For 50 µl sample of RNA: 1. Add 5 µl 10X Turbo DNase Buffer to the RNA and mix gently. 2. Add 1.5 µl DNase and mix gently. Incubate for at 37 ° C for 30 minutes (in shaker). 3. Add 1.5 µl more enzyme, and repeat incubation. 3. (Always resuspend reagent by flicking/vortexing tube before dispensing). Add 10 µl DNase inactivating reagent and mix well. 4. Incubate 5 min. at room temperature, mixing occasionally (2-3 times) to redisperse the DNase Inactivation Reagent. (Make sure room is between 22-26° C or can decrease activity of this step, leaving DNase in the reaction). 5. Centrifuge at 10,000 x g for 1.5 minute and carefully transfer the RNA (supernatant) to a fresh tube (be sure to avoid the pelleted inactivation reagent. 3 C. cDNA synthesis. As per Affymetrix expression_analysis _technical_manual.pdf, except using Superscript III and inactivating this enzyme for 15 minutes at 70 C. Dilute poly-A RNA controls (Affy #900433) 1. Make 2 serial 1:20 dilutions of stock RNAs (for use with the 100 (midi) Format Medicago chip) 2. Add 2 μl in each 30 l reaction below Allow a minimum of 30 µg of RNA per chip (Barnett et al. 2004). Perform reactions in triplicate, using 10 µg of RNA in each. cDNA yields may be less 3. Prepare the following mixture in new PCR strip-tubes: Primer Hybridization Mix Components Volume/amount Final concentration Total RNA 10 µg 0.33 µg Random 10 µl 25 ng/µl primers Hexamers (75 ng/µl) (Invitrogen) Diluted polyA RNA controls (Affymetrix #900433) Nucleasefree H2O 2 µl Total volume 30 µl variable Up to 30 µl 4. Using a thermal cycler, incubate the RNA-Primer mix at 70°C for 10 min followed by 25°C for 10 min and then chill to 4°C. 5. Prepare triplicates of the reaction mix for cDNA synthesis and add to the annealed RNA-primer mix. 4 cDNA Synthesis Reaction Mix Components Volume/amount 5X first strand buffer 12 µl (Invitrogen) 100 mM DTT 6 µl (Invitrogen) 10 mM dNTPs 3 µl (Invitrogen) SUPERase Inhibitor 1.5 µl (20 unit/µl) (Ambion, Austin TX) SuperScript III 7.5 µl (200 unit/µl) (Invitrogen) Annealed RNA-primer mix 30 µl Total volume 60 µl Final concentration 1X 10 mM 0.5 mM 0.5 unit/µl 25 unit/µl 6. Incubate the reaction at 25°C for 10 min, 37°C for 60, then 42°C for 60 min. 7. Inactivate the enzyme at 70°C for 15 min and hold at 4°C. Removal of RNA 8. Add 20 µl of 1 N NaOH and incubate at 65°C for 30 min. 9. Add 20 µl of 1 N HCl to neutralize. 5 D. Purification and Quantification of cDNA Synthesis Products (Modified from Qiagen QIAquick PCR Purification Kit protocol. Qiaquick spin columns are the same in Qiagen gel extraction or PCR purification kits) All spins at 12000 rpm (~15000 g) in an Eppendorf microfuge: 1. Add 500 μl of Buffer PB to cDNA sample (100 μl) and mix. Make sure to use binding buffer PB; avoid any buffer with pH-indicator dye, which will interfere with microarray reactions. 2. Put sample on Qiaquick spin column (in collection tube) and spin for 60 seconds. 3. Discard flow-through and put column back in the same tube. 4. To wash, add 750 μl Buffer PE to column and spin 60 seconds. 5. Discard flow-through and place column back in the same tube. Spin again to remove residual ethanol. 6. Place column in clean microcentrifuge tube. 7. Add 20 μl H20 of pH>~7.0. Let stand for 60 seconds, then spin for 60 seconds to elute DNA. 8. Repeat step 7 with same collection/microfuge tube (~40 μl elution volume total). 9. Leave tubes, caps open, on bench top at room temp for 20 minutes to remove residual ethanol. Quantify the purified cDNA product by 260-nm absorbance (1.0 A260 unit = 33 µg/ml of single-strand DNA). For a microcuvette use 1 μl in 50 μl water. Visualize cDNA on agarose gel (see Figure 2) 6 E. cDNA Fragmentation cDNA concentrations were adjusted for use with our GeneChip. Terminal labeling was doubled for root nodules and plant roots to ensure excess label for these reactions (Barnett et al. 2004). Use total of 12 µg cDNA for root nodule chips. 1. Prepare the following reaction mix: Fragmentation Reaction: Components Volume/amount Concentration 10X One-Phor-All buffer 5 µl 1X (USB part 78331, ask for Affymetrix price) cDNA 40 µl 12 µg DNAse I (see note) x µl x unit/µg of cDNA Nuclease-free H2O Up to 50 µl Total volume 50 µl Note: The amount of DNase I depends on its titer which must be checked (see below). Dilute DNase I to x unit/µl in 1X One Phor-All-Buffer. Use immediately and do not store. 2. Incubate the reaction at 37°C for 10 min. Inactivate the enzyme at 98°C for 10 min. 3. Add fragmented cDNA directly to the terminal labeling reaction. Alternatively, the cDNA can be stored at -20°C and labeled later. 4. To examine fragmented cDNA, load 200 ng on a 1.5% agarose gel using an Orange G load buffer and stain with sybr GOLD (details are given in Affymetrix manual). Ethidium bromide may be used but will give very faint fluorescence with singlestranded DNA (see Table 2 and Figure 3 below). 7 The remainder of the protocol was performed by Thanh Nguyen at the University of Toronto CAGEF processing facility, following the Affymetrix protocol for 100 Format (Midi) GeneChips with double terminal labeling and a hybridization temperature chosen to accommodate plant and bacterial genomes (Barnett et al 2004). Details of the hybridization and wash protocols are specific to the DNAs and the CAGEF Facility. See Affymetrix manual for suggestions. Terminal Labeling Reagents: GeneChip DNA Labeling Reagent, Affymetrix p/n 900542 – to label the 3’termini of the fragmentation products. Terminal Deoxynucleotidyl Transferase, Recombinant, Promega# M1871 including - Terminal Deoxynucleotidyl Transferase, Recombinant, and - Terminal Transferase 5X Buffer 1. Prepare the following Reaction Mix: edited from Section 3. Prokaryotic Sample and Array Processing / Terminal Labeling Table 3.1.5 = terminal Label Reaction (page 3.1.12) Terminal Label Reaction 2X (Nodule and Plant) Components x1 (μl) cDNA = 49μl sample 1-8 x9 5x Reaction Buffer GeneChip DNA Labeling Reagent, 7.5mM Terminal Deoxynucleotidyl Transferase Fragmentation cDNA product (nodule = 12ug) Water 16.00 4.00 4.00 47.75 8.25 16.00 4.00 4.00 49.00 7.00 144 36 36 Total Volume = 80μl 80.00 80.00 279 take 31μl 63 2. Incubate the reaction @ 37oC for 60minutes. 3. Stop the reaction by adding 2μl of 0.5M EDTA. Alternatively, the reaction mixture may be stored @-20oC. 8 Prokaryotic Target Hybridization 1. Prepare the following solution mix: Hybridization Cocktail see table in the GeneChip Expression 3’Amplification The reason to scale down the volume of the hybridization cocktail to 210μl is to save on the amount of cDNA input - due to the original lowcDNA yield. 2X Hybridization Mix 3nM B2 Control 20X Control (65oC) DMSO Fragmented + Labeled cRNA Water Total Volume = 210μl 1 105 3.5 10.5 21 70 210 x9 945 31.5 94.5 189 1260 take 140μl per sample Note: 2. 3. 4. 5. Heat 20x control at 65 C for 5 mins before adding to the cocktail Protocol could be stopped here. Store samples @ -20oC. Equilibrate probe array to RT immediately before use. Add 200μl of the Hybridization Cocktail to the probe array. Cover the septa with Tough Spots Hybridize @ 48oC, 60rpm, for 16h. in an Affymetrix Hybridization Oven 640. 9 Probe Array Washing and Staining -prime the Fluidics Station -the next day, transfer hybridization samples from gene chipslabeled microtubes store -20oC -fill Chips with 250μl of WashA – to prevent drying Using Hybridization +Wash+Stain Kit pn. 900720 -aliquot 600μl of Stain1 for tube 1 - aliquot 600μl of Stain2 for tube 2 -aliquot 800μl of Array Holding Buffer for tube 3. Fluidics Scripts: -use FS450_001 for Medicago Chip In case of bubble in the chip after staining, 800μl of the Wash A could be used to refill tube 3. 10 DNase I Titer Each lot no. of DNase I can vary greatly. To titer DNase I, determine the amount of DNase I needed to fragment 1 µg of cDNA to sizes between 50 and 200 bases. Choose between 0.2 and 1.1 DNase I units/µg cDNA to start. Prepare the reaction mix as above with less cDNA to start: Components Volume/amount Concentration 10X One-Phor-All buffer 5 µl 1X (USB part 78331, ask for Affymetrix price) cDNA 40 µl 3-7 µg DNAse I (see note) x µl x unit/µg of cDNA Nuclease-free H2O Up to 50 µl Total volume 50 µl TRIAL RUN TITRATION REACTION: For initial chip, titer DNAse using 5 reactions (range from 0.2 – 1.1 Units / µg): Components Volume/amount Concentration 10X One-Phor-All buffer 0.72 µl 1X (USB part 78331, ask for Affymetrix price) cDNA 5.78 µl 1 µg DNAse I (see note) x µl x unit/µg of cDNA Nuclease-free H2O Up to 7.6 µl Total volume 7.6 µl DNAse titration reactions. Gel stained with ethidium bromide, BPB and Orange G stains partially obscure fluorescence 100bp cDNA units / µg cDNA 0.2 0.6 0 .8 1.1 100bp 0.2 units / µg cDNA 0.4 0.6 0.8 1.1 11 Tables and Figures Table 1. RNA extractions of nodule tissue. 2 plants from each treatment combination were pooled in a single RNA extraction. Plant Treatment Block Plant Rhizobium Date Sample genotype genotype extracted 3/124 A 1 Naut1 SalsB 37/75 B 1 Naut1 SalsC cDNA on chip 5/25/2009 bot1446 10.2 ug 5/25/2009 bot1447 10.5 81/116 C 18/62 D 1 1 Sals4 Sals4 SalsB SalsC 5/25/2009 bot1448 11.1 5/25/2009 bot1449 8.5 Plant Treatment Block Plant Rhizobium Date Sample genotype genotype extracted Chip name 10/67 A 79/125 B 2 2 Naut1 Naut1 SalsB SalsC cDNA on chip 6/15/2009 Bot1489 7.3 6/15/2009 Bot1490 6.6 26/47 19/68 2 2 Sals4 Sals4 SalsB SalsC 6/15/2009 Bot1491 4 6/15/2009 Bot1492 6.1 C D Plant Treatment Block Plant Rhizobium Date Sample genotype genotype extracted Chip name Chip Name 25/80 11/58 A B 3 3 Naut1 Naut1 SalsB SalsC cDNA on chip 9/11/2009 Bot1493 8.2 9/11/2009 Bot1496 6.0 4/52 5/69 C D 3 3 Sals4 Sals4 SalsB SalsC 9/11/2009 Bot1495 9.4 9/11/2009 Bot1494 10.2 12 Table 2: Example of fragmentation reaction for sample 146. Components 10X One-Phor-All buffer (USB part 78331, ask for Affymetrix price) cDNA DNAse I (see note) Nuclease-free H2O Total volume Volume/amount 5 µl Concentration 1X 44 µl (0.75 µl (10 unit / µl)) Up to 50 µl 50 µl 12.5 µg (0.6 unit/µg of cDNA) 13 Figure 1: Example gel of total RNAs. RNAs were loaded on a 1.5 agarose 0.5x TBE gel and stained with ethidium bromide. Bands for the 4 rRNAs are visible for all samples. 14 Figure 2: Visualization of cDNA on agarose EtBr gel. Ran 3 µl cDNA + 2 µl loading buffer. 100 bp 146_1 146_2 162_1 15 Figure 3. Results of fragmentation reaction for sample 146. 2 µl each of cDNA and fragmentation reaction in Orange G load buffer were run on a 1.5% agarose, 0.5x TBE gel and stained with ethidium bromide. The fragmented DNA-ethidium bromide complex was faint but clearly visible, and is just visible in the photograph. 1 2 3 100 bp cDNA frag 16