Biotechnology Final Report
advertisement

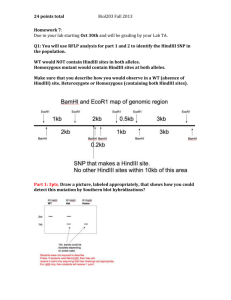
Josh Hunt 5/2/24 Biotechnology Lab Plasmid Identification Project In our class, we were assigned to incubate and identify our given set of DNA. Using techniques taught in the B TECH 1015 curriculum, we’ve been able to get to the point of successfully working with plasmids in order to get desired results. To do this, I needed to created a proper plasmid. A plasmid is a small section of DNA that has been removed from its chromosomal DNA within the cell. Within this plasmid, restriction enzymes are added in order to cut the DNA at certain spots the enzymes recognize. I then ran these substances through gel electrophoresis. This enables those studying DNA to measure the basepairs of the sample they’re studying. The measurements of these base pairs are able to predicted at sources like on the the website NEBCutter. These predictions enable people studying DNA like me, to match our data with correlating predictions from the website. In this way, I am able to predict the plasmid I have been given from 3 possible plasmids. After thorough experimentation, I’ve concluded that the plasmid I have been given is pKAN. Having been given an enzyme with the code name 6025C19, I first decided upon the recipe to use for the digest. Having been given a concentration from my teacher, I determined to use a specific amount of plasmid for my recipe. We obtained both our marker DNA, our restriction enzymes, and our reaction buffer from NEB, a science company that stands for New England Biolabs. The next step was to then determine how much and what kind of restriction enzymes I would use. After conducting some research, I found the enzymes BglI, PstI, and BamHI to use for my incubation. I decided to use BglI for one trail, and PstI and BamHI for another. This was due to the idea that each of these enzymes cut the plasmids at least 2 or more times. In my second trial, I used a fourth enzyme, HindIII, which I felt would give more accurate data. I then determined the buffer necessary in order to allow proper incubation, which I found at NEB’s website. After finding this, I was able to determine the amount of dH2O to add. After assembling these, I allowed my digests to sit for about an hour and a half at 37 oC. This was perhaps, in my opinion the most important part of the whole procedure, as this determined whether or not the enzymes incubated the DNA. In my first experiment, my BglI digest did not incubate, and in my second experiment, I found my BglI, as well as my HindIII digests, in not fully incubate either. One conclusion drawn from this is that these restriction enzymes need more time, or perhaps, a higher temperature, to incubate. Using a .8% concentration of agarose, I added my determined amount of agarose. I then measured out the amount of x10 TAE in a graduated cylinder, filling the rest of the amount with water. I then transferred the contents to the flask containing the agarose. After microwaving this for about a minute and 20 seconds, I let my solution cool, and then poured it into the gel tray. I then ran my gel, which was at about 140 volts the first time, and 130 volts the second time. The time these ran were about 40 to 50 minutes each, by the time they reached the 4.5 mark. I used a x10 TAE buffer, using a tenth of 280 mL, which was the total volume. After my enzymes were ready, I used a UV image system to take the picture of my gel. I found that only my PstI+BamHI was the only digest I ran that actually incubated. From this, I was able to determine my fragments. To do this, I first matched the ladders together, having 10 ladders the first time (with a 0.5 kbp), and the second time which did not have the 0.5 kbp marker. I ran a line through each of the lines shown in the digests that incubated. I then created a line of best fit on a graph using the ladders. From that, using the equation excel gave me, I was able to find the fragment sizes of the digest sections. My data correlated very nicely with my predictions. However, after checking my predictions again, I found I had put the settings on linear instead of circular. After correcting my mistake, I found that my data did not work predictions. with the However, after consulting my teacher, I was advised to consider if one of my cuts never actually happened. Thus, I allowed to was determine my having the plasmid. Figure 1 After concentration being told to me, which was 250 µg/mL, I found my total miniprep of plasmid to be about 2 µL. Figure 1 shows my first trial, when I did not include HindIII. F my second trial, in which neither my HindIII nor my BglI Figure 2 digests incubated. However, I was able to determine the plasmid by looking at the data given me by my PstI + BamHI incubation. When I plugged my data into excel, I found highly positive results. I was able to find that my measurements were very accurate. My DNA Figure 3 Figure 4 fit nicely along the exponential line. Trial One Travel Distance (mm) Base Pairs Difference from Predictions PstI+BamHI #1 36.5 3462.7 -191.696 PstI+BamHI #2 48.6 1045.14 -122.137 Trial Travel Distance (mm) Base Pairs Difference from Predictions BglI #1 30.1 4113.6 80.4019 PstI+BamHI #1 31.9 3386.84 -115.843 PstI+BamHI #2 43.7 946.957 -23.9567 Predictions BglI PstI+BamHI HindIII pAMP 3263, 1118, 158 4468, 71 4539 pKAN 3139, 794, 261 2870, 923, 401 4194 pBLU 2121, 1710, 1576 3902, 1316, 197, 22 5437 pKAN (without second cut) According to my trials, 3271, 923 I am anywhere from 200 to 1000 base pairs off from what I’m supposed to be at, according to me predictions. However, after consulting my teacher, I was able to determine that in my pKAN PstI+BamHI solution did not cut at the second cut, thus I got a good fit that is +200 basepairs off. From that data, I can safely conclude that my plasmid is pKAN, as my results match up with the base pairs. A possible explanation as to why things happened the way they did is within the incubation process. According to my knowledge, I would hypothesize that PstI does not take a long time to incubate. On the other hand, BamHI does, which would explain why my results were the way they were. References Nucleotide Sequences of Plasmids Nucleotide Sequences of pAMP, pKAN, & pBLU Plasmids. Internet: http://www.dnalc.org/resources/plasmids.html NEBcutter V2.0 o NEBcutter V2.0. Internet: http://tools.neb.com/NEBcutter2/ Buffer Finder o Double Digest Finder. Internet: https://www.neb.com/tools-andresources/interactive-tools/double-digest-finder Mr. Scott o Scott, R. (2014, May 2). o