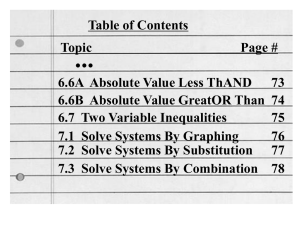
click here
advertisement

1. Each base pair in B form DNA is separated by 3.4 Ao (1 Ao = 10-8 cm) (3.2 x 106 bp) (3.4 x 10-8 cm/bp) = 1.08 x 10-2 cm x 104 2. B form DNA completes 1 turn (revolution) every 10 bp. (3.2 x 106 bp)/ 10 bp/revolution = 3.2 x 105 revolutions/ 60 min = 5,333 rpm/2 replication forks = 2,700 rpm/fork (c) 3. BamHI is a six base cutter. The odds of any base at a given position is 1/4, so we would expect the odds of any random six bases to be (1/4)6 = 1/4096 or 1 site/4096 bp 3.2 x 106 bp/ 4096 bp/site = 780 sites (b) 4. The recognition sequence is CPyCGPuG. For positions 1,3,4 and 6 in this sequence only 1 base out of four will lead to cutting. For positions 2 and 5 in the sequence, two bases out of 4 will lead to cutting. Therefore, the odds of having this exact sequence in a random DNA molecule will be: 1/4 x1/2 x 1/4 x 1/4 x 1/2 x 1/4 = 1/1024; or it will cut once every 1024 base pairs. Ans: 1024 bp (c) 5. The results of separating the two strands of DNA will leave a 4 base overhang…this 4 base overhang happens to be THE SAME for the enzymes BamHI and BglII: 5’...G GATCC...3’ 3’...CCTAG 5’...T CTAGA...3’ 3’...AGATC 5’...A G...5’ T...5’ GATCT...3’ 3’...TCTAG A...5’ Therefore, the single strand overhangs for these two enzymes would allow them to base pair with each other. Please note, however, that if we used these two enzymes for cloning, we would not reconstitute either a BamHI or BglII site after ligation: e.g. 5’…AGATCC…3’ 3’…TCTAGG…5’ Ans: (c) 6. Gln-Tyr-Met-Phe-Asp-Glu-Trp. Ans (c) 7. 2 x 2 x 1 x 2 x 2 x 2 x1 = 32 Ans (d) 8. Ans: (b) False, it CAN NOT be used for Northern blots. An mRNA would encode the same sequence, and the probe would NOT be complementary to it- it would be the same sequence. 9. Both the 8 and 2 kb fragments (c). + + X + D 10. Two cross-overs are observed in this data set (II-5, II-14, above)…2/14 = 0.143 = 14.3 m.u. (d).