Additional file 1

Additional file 1
99.7% MnP3 PC15-1089546
MnP3 PC9-137740
MnP5 PC15-199511
99.7% MnP5 PC9-137765
VP1 PC9-137757 *
VP2 PC15-1113241
99.1% VP2 PC9-137766
100% MnP1/VP PC15-1096331
MnP1/VP PC9-137760
VP3 PC15-156336
99.7% VP3 PC9-123383
100% MnP4 PC15-1099081
MnP4 PC9-121638
100% MnP6 PC15-1041740
MnP6 PC9-51713
MnP2 PC15-199510
100%
MnP2 PC9-137764
98.5%
99.0%
DyP4 PC15-1069077
DyP4 PC9-117204
DyP1 PC15-62271
99.8%
98.7%
99.7%
99.7%
DyP1 PC9-87639
DyP2 PC15-1092668
DyP2 PC9-115057
DyP3 PC15-52170
DyP3 PC9-97865
CCP PC15-1096819
CCP PC9-77045
98.8%
HTP1 PC15-1110336
HTP1 PC9-114464
HTP3 PC15-1108212
HTP3 PC9-127284
HTP2 PC15-1111884
99.2
HTP2 PC9-123372 class-II (POD)
DyP class-I
HTP
1.2
1.1
1.0
0.9
0.8
0.7
0.6
0.5
0.4
0.3
0.2
0.1
0.0
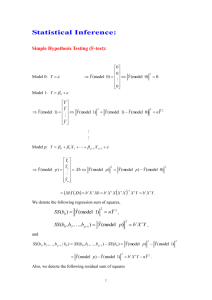
Figure S1. Phylogram of heme peroxidase sequences from the genomes of two P. ostreatus monokaryons.
The evolutionary analysis of the deduced peroxidase sequences was performed with
MEGA5 using Poisson distances and UPGMA clustering. Accession numbers correspond to the monokaryons PC9 v1.0 and PC15 v2.0 catalogs. The sequence identities between each pair of allelic models are shown (*indicates the existence of a premature termination codon in PC15
1089895, resulting in a pseudogene not included in the phylogram). Three main clusters were identified corresponding to POD (VP and MnP), DyP and HTP sequences, together with a unique class I gene (CCP). The 1096331/137760 PC15/PC9 models, initially classified as VPs [1], are reclassified as MnP1.
1
Figure S2. Amino-acid composition of the nine VP and MnP isoenzymes from the P. ostreatus genome (predicted mature proteins, Figure 1).
The comparatively high number of lysine residues
(20 compared with the 10 average) in MnP4 is among the most significant differences observed.
2
kDa
75
50
37
A
1 2 3 4 5 6 7
25
B
MnP3 VP2 MnP1
Figure S3. E. coli expression of P. ostreatus genome peroxidases (SDS-PAGE).
( A) Time-course of MnP4 overexpression in IPTG-induced cultures (lane 1, standards; lane 2, purified MnP4; lanes
3-7, total proteins 0, 1, 2, 3 and 4 h after induction, respectively). ( B) Inclusion bodies from MnP3,
VP2 and MnP1 isoenzymes.
3
A
B
C pH pH
D
E
F pH pH
I
G
H pH pH pH pH
Figure S4.
pH 2-9 stability of the nine PODs from the P. ostreatus genome at different incubation times.
Residual activities after 1 min (green), 1 h (magenta), 4 h (orange) 24 h (blue) and 120 h (purple) incubation at 4 ºC in 100 mM B&R buffer (pH 2-9) were estimated for VP1
(A) , VP2 (B) ,
VP3 (C) , MnP1 (D) , MnP2 (E) , MnP3 (F) , MnP4 (G) , MnP5 (H) , and MnP6 (I) . Activity was assayed as described in Figure 2 . Means and 95% confidence limits.
4
A
B pH pH
Figure S5. Influence of temperature on the pH stability of PODs from the P. ostreatus genome.
The stable MnP4 (A) and the unstable MnP3 (B) were incubated for 4 h at 4 ºC (dark-blue bars) or
25 ºC (red bars) in 0.1 mM B&R buffer (pH 2-9) and the residual activity was estimated as described in Figure 2 . Means and 95% confidence limits.
5
A
B
C
D
Figure S6. Stereo views of some of the main differences between VP1 (light blue ) and MnP4
(light brown) crystal structures. (A) Loop close to the distal Ca 2+ ion ( Figure 5A ). (B) Main heme access channel region ( Figure 5C ). (C and D) Two sequence stretches (V248-P252 and P286-H293 in VP1, and I254-S259 and R292-P298 in MnP4) at the back of the protein, from two different orientations (the heme cofactor, proximal Ca 2+ and its ligands, and VP1 W164 are also visible)
( Figure 5B ).
6
Figure S7. Partial 2Fo-Fc electron density map, contoured at the 1.1 σ level, of heme cofactor, some neighbor residues and several water molecules in the VP1 and MnP4 crystals. (A and C)
Stereo views of VP1 and MnP4, respectively, from the heme δ-position side. The heme pocket is delimited by H39, E40, L42, R43, F46, H47, P139, P141, I148, M152, V162, L165, L166, S168,
H169, I171, A172, A173, A174, D175, K176, V177, F186, L228, S230, L234, M262 and M265 in
VP1, and H39, E40, L42, R43, F46, H47, P145, P147, I154, M158, V168, L171, L172, S174,
H175, V177, A178, A179, Q180, D181, T182, I183, F192, L234, S236, L240, M268 and M271 in
MnP4, some of them being shown in the stereo views. (B and D) Detail of the electron density maps of several water molecules in MnP4 and VP1, respectively, from the heme α-position side, also showing two conserved histidine and one conserved arginine residues.
7
120
100
A
80
60
40
20
0
1 10 100
ABTS (µm)
1000 10000
0,8
0,6
0,4
B
0,07
0,06
0,05
0,04
0,03
0,02
0,2
0,01
0,0
0,0 0,1 0,2 0,3 0,4
0,00
1/[ABTS] (µm -1 )
Figure S8. Sigmoidal curve for ABTS oxidation by VP isoenzyme VP1 (A) enabling calculation of two sets of kinetic constants (B) as shown in Table 1 .
1. Ruiz-Dueñas FJ, Fernández E, Martínez MJ, Martínez AT: Pleurotus ostreatus heme peroxidases: An in silico analysis from the genome sequence to the enzyme molecular structure.
C R Biol 2011, 334: 795-805.
8