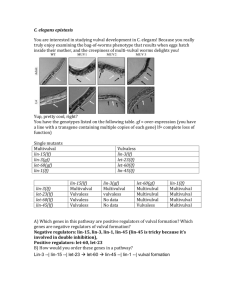
1.) Each cell in the Drosophila wing produces a hair that points
advertisement

1.) Each cell in the Drosophila wing produces a hair that points towards the tip of the wing. Researchers wondered how each cell knew which way the wing tip was. Screens were carried out and mutants identified where the consistent hair orientation is disrupted. You obtain one of the mutant strains which contains a mutation in frizzled and decide to characterize it further. Wildtype PCP mutant A: You first decide to ask whether frizzled is acting autonomously or nonautonomously to coordinate hair orientation. What technique will you use? Ans: mosaic analysis B: What is the genotype of the fly that you will analyze? Ans: wing driver: flp/+; FRT fz/FRT tub:GFP C: How will you obtain this fly? Assume you have three starting strains consisting of 1) engrailed::FLP (on the second chromosome) 2) FRT, fz15 (on the third chromosome) and 3) FRT tubulin:GFP. What fraction of the progeny will be what you want? Cross1 en:FLP x FRT tub:GFP Cross 2 en:FLP/+; FRT tub:GFP/+ x FRT, fz ¼ of progeny will be right can be identified by looking for GFP- clones in the wing. D: What do you expect to see if frizzled is cell autonomous or non autonomous? Note in this example the outlined clone will be dark, and the surrounding cells will be GFP+. If the gene acts in a purely cell autonomous fashion, then the mutant phenotype is restricted to the clone. E: Talking to your lab mates you realize that lots of them are going to be doing the same experiment you just did only using their favorite genes. As a public service you decide to make the engrailed:FLP; FRT tubulin-GFP fly line. How will you go about doing this? For this problem you cannot distinguish by fluorescence one copy vs two of GFP and the engrailed: FLP is unmarked. Explain at every step how you will select the flies you want. Hint: your lab has a L/Cyo; TM3/TM6 double balanced stock. Ans En:FLP x L/Cyo; TM3/TM6 select non lobed progeny En:FLP/Cyo; TM3/+ x L/Cyo; TM3/TM6 select non lobed both TM3 and TM6 progeny En: FLP/Cyo; TM3/TM6 x L/Cyo; FRT tub:GFP/TM3(these flies obtained from basically the same series of crosses only using the FRT line) select non lobed, GFP positive w/ TM6 marker flies En:FLP/Cyo; FRT tub:GFP/TM6 x to itself; select progeny w/o balancers 2.) You are an ambitious young fly geneticist studying the reproductive system. After feeding your flies plenty of nasty chemicals, you create balanced stocks of two interesting mutations. Neither mutations are lethal, but both result in adult female sterility. Upon sequencing your mutants, you find truncations in two different genes (Neg1 and Neg2, named for no viable eggs) that show homology to mammalian genes involved in gonad formation. A)First, you want to look at Neg1 + 2 protein expression in a wildtype fly. How would you do this? (Assume that you are an expert at molecular biology, so shuffling promoters and creating transgenic flies is a cake walk.) There are many possibilities, but be sure to explain the advantages / disadvantages to any method you choose. Many options: Pneg (= promoter region of Neg gene) (driving) GFP o Are you sure you get ALL of the regulatory elements? Maybe you will see aberrant expression Pneg Gal4, UAS-GFP o You still may miss some regulatory elements o You may be able to use the Gal4 to drive something else later o UAS-GFP / RFP / YFP are readily available Neg-GFP fusion protein at endogenous locus o More reliable expression pattern o Fusion may interfere with function Antibody stain o Unlikely that a drosophila antibody and protocol exists for your new mutant B) Your first experiments show Neg2 to be expressed in the posterior part of the adult germarium (small, reproductive unit of the fly ovary) but you don’t see any Neg1 expression. You wonder if Neg1 is expressed transiently during development. Design a fly that would allow you to label cells that transiently express Neg1. Pneg1 FLP, Tub-FRT-stop-FRT-GFP Note that not all cells labeled will have expressed Neg1 at some point. You see all cells that are descended from a transiently-Neg1-expressing cell, even if the individual cells have never expressed the protein. C.) You have noticed that Neg1 and Neg2 result in no cell polarization in the epithelial cells surrounding the egg. You are interested in determining if your phenotype is cell autonomous or non-cell autonomous, so you want to make negatively marked clones. What two strains would you need in order to do this experiment? Strain 1: hsFLP/ hsFLP ; Neg1[mut], FRT/ cyo; Strain 2: tub-gal4,UAS::lacZ(or UAS::GFP), FRT/ cyo D.) For the two chromosomes undergoing recombination draw out what you would expect to see. E.) But, looking for absence of your reporter is annoying. What construct could you make instead that would generate positively marked clones? Why is this sometimes easier? For small clones, this is easier to identify. hsFLP; Neg1[mut], FRT / tub-gal80, FRT; UAS-GFP / tub-Gal4 This is equivalent to the MARCM technique for generating positively marked clones. F.) Through careful BLAST searching, you discover that Drosophila has a well conserved ortholog of a mouse gene that is important for germline development known as Nubn3. You order a line that contains a P-element insertion that is predicted to disrupt dNubn3. The line comes as a balanced stock (P[w+]Nubn3/CyO). You cross siblings and recover no non Cy flies. You conclude that dNubn3 is an essential gene, and the P-element insertion is recessive lethal. Your PI says that the proper way to do this is to examine the phenotype of flies with the insertion mutation over a chromosome with a deletion of 15 kb around, and including dNubn3. Why is this the better method? When you cross P[w+]/CyO with itself, you’ve homozygoused not only the insertion, but the entire chromosome. It’s possible that the lethality is due not to the insertion in dNubn3, but to some other mutation on the chromosome (remember it’s a balanced stock, so the chromosome can accumulate additional recessive lethal mutations without a selective disadvantage). This is especially true for mutations generated by p-element mobilization. Examining the transheterozygote (P[w+]nubn3/Def) is a more convincing demonstration that the lethality is due to loss of Nubn3 function. The assumption here is that the two different null alleles have different genetic histories, so they are unlikely to share the same linked lethal. G.) The transheterozygote (P[w+]Nubn3/Def) is still dead, the flies die as larvae. You're still interested in determining if dNubn3 plays a role in egg production in the adult female germline. How could you test this? Use the GAL4/UAS system to drive Nubn3 RNAi specifically in the female germline. Essentially combining UAS-shRNA (Nubn3) with a female germline driven Gal4. Or you could use mosaic analysis to induce mutant clones in the female germline. Generate cells homozygous for the p-element insertion, in a heterozygous background. You would need to do this early to allow the mutant cells to populate most of the germline. Maybe even use the minute technique to make them faster growing than the competing surrounding heterozygous cells. H.) Your analysis reveals that dNubn3 is required for a functional female germline. Why do we need to be a little careful about over interpreting this result? There could be a problem with the specificity of the phenotype. We’ve essentially forced specificity by driving the RNAi/inducing mutant clones only in the female germline. We need to be careful about what we say about the actual biology of Nubn3 in the whole organism. For example driving RNAi against a ribosomal protein in the germline would also give you a female germline phenotype, even though translation has nothing specifically to do with germline biology.