Supplement 1
advertisement
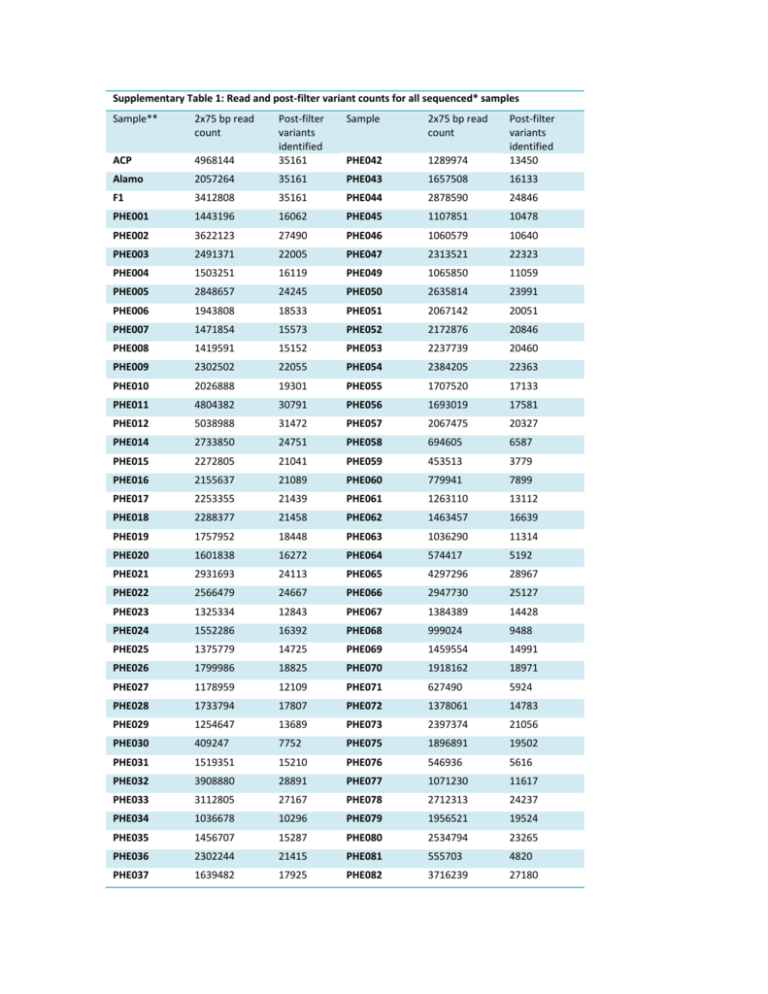
Supplementary Table 1: Read and post-filter variant counts for all sequenced* samples Sample** 2x75 bp read count Sample 2x75 bp read count 4968144 Post-filter variants identified 35161 PHE042 1289974 Post-filter variants identified 13450 ACP Alamo 2057264 35161 PHE043 1657508 16133 F1 3412808 35161 PHE044 2878590 24846 PHE001 1443196 16062 PHE045 1107851 10478 PHE002 3622123 27490 PHE046 1060579 10640 PHE003 2491371 22005 PHE047 2313521 22323 PHE004 1503251 16119 PHE049 1065850 11059 PHE005 2848657 24245 PHE050 2635814 23991 PHE006 1943808 18533 PHE051 2067142 20051 PHE007 1471854 15573 PHE052 2172876 20846 PHE008 1419591 15152 PHE053 2237739 20460 PHE009 2302502 22055 PHE054 2384205 22363 PHE010 2026888 19301 PHE055 1707520 17133 PHE011 4804382 30791 PHE056 1693019 17581 PHE012 5038988 31472 PHE057 2067475 20327 PHE014 2733850 24751 PHE058 694605 6587 PHE015 2272805 21041 PHE059 453513 3779 PHE016 2155637 21089 PHE060 779941 7899 PHE017 2253355 21439 PHE061 1263110 13112 PHE018 2288377 21458 PHE062 1463457 16639 PHE019 1757952 18448 PHE063 1036290 11314 PHE020 1601838 16272 PHE064 574417 5192 PHE021 2931693 24113 PHE065 4297296 28967 PHE022 2566479 24667 PHE066 2947730 25127 PHE023 1325334 12843 PHE067 1384389 14428 PHE024 1552286 16392 PHE068 999024 9488 PHE025 1375779 14725 PHE069 1459554 14991 PHE026 1799986 18825 PHE070 1918162 18971 PHE027 1178959 12109 PHE071 627490 5924 PHE028 1733794 17807 PHE072 1378061 14783 PHE029 1254647 13689 PHE073 2397374 21056 PHE030 409247 7752 PHE075 1896891 19502 PHE031 1519351 15210 PHE076 546936 5616 PHE032 3908880 28891 PHE077 1071230 11617 PHE033 3112805 27167 PHE078 2712313 24237 PHE034 1036678 10296 PHE079 1956521 19524 PHE035 1456707 15287 PHE080 2534794 23265 PHE036 2302244 21415 PHE081 555703 4820 PHE037 1639482 17925 PHE082 3716239 27180 PHE038 1690629 18494 PHE083 870613 9305 PHE039 1862999 15212 PHE084 1033877 11839 PHE040 1620897 17103 PHE085 1772703 18172 PHE041 2373533 22650 PHE086 4800130 30899 * PHE013, PHE48, and PHE74 died during transplanting. ** PHE is the identifier for F1BC1 samples. M 1 2 3 4 5 6 7 8 9 10 11 12 21.2 kb 5.1 kb 4.3 kb 3.5 kb 2.0 kb 1.9 kb 1.6 kb 1.4 kb 0.83 kb 0.56 kb Supplementary Figure 1: Southern blot of bar transgene amongst 3% Finale resistant and sensitive switchgrass. A single transgene insertion event is identified in the transgenic Alamo parent, F1, and HbR F1BC1. The transgene was not identified in any HbS individual. 1) T85-2 Alamo paternal parent, 2) ACP maternal parent, 3) Wild-type Alamo 4) F1, 5) HbS F1BC1-1, 6) HbS F1BC1-2, 7) HbS F1BC1-3, 8) HbS F1BC1-4, 9) HbR F1BC1-1, 10) HbR F1BC1-2, 11) HbR F1BC1-3, 12) HbR F1BC1-4. B 7 35 Inforescence Rachi Count 40 6 5 µg/ml Chl C 8 4 3 2 1 2000 1800 Inforescence Spikelet Count A 30 25 20 15 10 1600 1400 1200 1000 800 600 400 5 0 0 Alamo ACP F1 D 200 Alamo ACP Alamo ACP F1 Alamo ACP F1 F E 1400 600 0 F1 1000 900 1200 400 300 200 Abaxial Stomata Count 800 Adaxial Stomata Count Inforescence Seed Count 500 1000 800 600 400 700 600 500 400 300 200 100 200 100 0 0 Alamo ACP F1 0 Alamo ACP F1 Supplementary Figure 2) Phenotypic plant characteristics of Alamo and ACP parents and the F1 hybrid. A) Leaf chlorophyll content. B) Rachis count, C) spikelet count, D) and seed count per inflorescence. E) Adaxial and F) abaxial leaf stomata count. M 1 2 3 4 5 6 7 8 9 10 11 12 13 M 766 bp 500 bp 300 bp 150 bp 50 bp Supplementary Figure 3) Segregation of bar transgene amongst 3% Finale resistant and sensitive switchgrass. Lanes are 1) negative control 2) positive control (100 ng transformation construct) 3) T85-2 Alamo paternal parent, 4) ACP maternal parent, 5) F1, 6) HbS F1BC1-1, 7) HbS F1BC1-2, 8) HbS F1BC1-3, 9) HbS F1BC1-4, 10) HbR F1BC1-1, 11) HbR F1BC1-2, 12) HbR F1BC1-3, 13) HbR F1BC1-4. A M 1 2 3 4 5 6 7 8 B 600 bp 500 bp 400 bp 300 bp 600 bp 500 bp 400 bp 300 bp 200 bp 200 bp 100 bp 100 bp M 1 2 3 4 5 6 7 8 Supplementary Figure 4) Chloroplast 49 bp indel and bar transgene. A) 49 bp indel in the chloroplast tRNA-Leu gene. B) bar transgene. 1) ACP maternal parent, 2) T85-2 Alamo paternal parent, 3) F1 4) F1BC1 PHE-01, 5) F1BC1 PHE-02, 6) F1BC1 PHE-03, 7) F1BC1 PHE-04, 8) negative control. Secondary bands (>600 bp) in the bar transgene gel are due to extended exposure time and difficulty in amplification. The bar amplicon (202 bp) is supported by both the size and correlation between the gel and the Southern blot with regards to positive and negative samples. 16 Sample Count 14 12 10 8 6 4 1 0.9 0.95 0.8 0.85 0.7 0.75 0.6 0.65 0.5 0.55 0.4 0.45 0.3 0.35 0.2 0.25 0.1 0.15 0 0 0.05 2 ACP Fraction of Genome Supplementary Figure 5: ACP fraction in F1BC1 offspring. The ACP fraction of each F1BC1 offspring genome was determined and samples were binned in intervals of 0.025 between 0 and 1. The mean ACP fraction per genome was 31.62% (± 6.35% (SD)). This is slightly above the expected value of 25%, likely due in part to segregation distortion due to selection against the BAR allele in the Alamo parent of the F1. A secondary factor may be some miscalled or misplaced markers due to the preliminary state of the genome assembly, which is supported by regions of miscalled ACP homozygosity that cannot exist in an F1BC1.







