NF-YC8 - Springer Static Content Server
advertisement

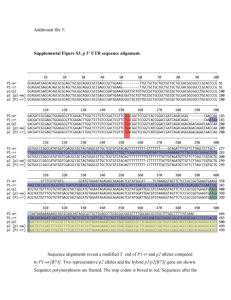
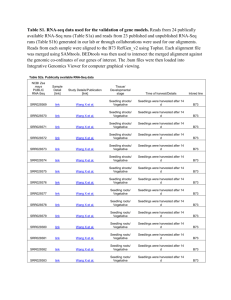
Mei et al. Supplementary Figures and Tables Figure S1 Phylogenetic analysis of NF-YC subfamily Figure S2 Maize NF-YC family multiple sequence alignments Figure S3 Sequence for CAPS analysis of NF-YC8 Figure S4 Sequence for methylation analysis of NF-YC8 5’ upstream region Figure S5 Sequence for methylation analysis of NF-YC8 coding region Figure S6 Sequence for methylation analysis of Mez1 Figure S7 Bisulfite sequencing for the DMR of Mez1 5’ upstream region Figure S8 Methylation analysis of the NF-YC8 5’ upstream region Figure S9 Methylation analysis of the NF-YC8 coding region Table S1 Methylated cytosine statistics of NF-YC8 in different alleles Table S2 Sequences of all PCR primers used Table S3 IDs for Arabidopsis thaliana NF-YC subfamily Mei et al. Figure S1 Phylogenetic analysis of NF-YC subfamily. Multiple sequence alignments were generated using ClustalX 1.83 on full-length proteins as implanted by MEGA 5.0 (Tamura et al. 2011; Thompson et al. 1997). Phylogenetic trees were constructed by neighbor joining (N-J) with the Possion correction and Pairwise deletion. Reliability values at each branch point represent the bootstraps cores (1,000 replicates) (Lin et al. Mei et al. 2011). Sequence of the mouse NF-YC subunit was used to root the tree. The functional annotation of Arabidopsis thaliana NF-YC was based on Petroni et al. (Petroni et al. 2012). (A) Phylogenetic tree of maize NF-YC subfamily. (B) Phylogenetic tree of maize and Arabidopsis thaliana NF-YC subfamily. Figure S2 Maize NF-YC family multiple sequence alignments. The secondary structures, alpha-helix (solid red cylinders) and coils (black lines), were represented on top of the amino acids sequence, predicted by PredictProtein and based on Liang et al. (Liang et al. 2014). The DNA-binding consisted of two amino acid AR and interaction domains were represented by black line. The region of dark blue outline shared with the structure of H2A (Liang et al. 2014). Mei et al. A B73 ACCAGGACCGACATTTACGACTTCTTGGTCGACATTGTTCCCAGGGATGAGATGAAGGACGACGGAATCGGGCTTCCTAG Mo17 ACCAGGACCGACATTTACGACTTCTTGGTCGACATTGTTCCCAGGGATGAGATGAAGGACGACGGAATCGGGCTTCCTAG Consensus accaggaccgacatttacgacttcttggtcgacattgttcccagggatgagatgaaggacgacggaatcgggcttcctag 80 80 B73 GCCCGGGCTGCCACCCATGGGAGCCCCAGCTGACGCATATCCATACTACTACATGCCACAGCAGCAGGTGCCTGGTCCTG Mo17 GCCCGGGCTGCCACCCATGGGAGCCCCAGCTGACGCATATCCATACTACTACATGCCACAGCAGCAGGTGCCTGGTCCTG Consensus gcccgggctgccacccatgggagccccagctgacgcatatccatactactacatgccacagcagcaggtgcctggtcctg 160 160 B73 GGATGGTTTATGGCGCCCAGCAAGGCCACCCGGTGACGTATCTGTGGCAGGATCCTCAGGAACAGCAGGAGCAAGCTCCT Mo17 GGATGGTTTATGGCGCCCAGCAAGGCCACCCGGTGACGTATCTGTGGCAGGATCCTCAGGAACAGCAGGAGCAAGCTCCC Consensus ggatggtttatggcgcccagcaaggccacccggtgacgtatctgtggcaggatcctcaggaacagcaggagcaagctcc 240 240 B73 GAAGAGCAGCAGTCTCTGCATGAAAGGGACTGAGGATGTCGCTCA..AGCTATCACCTGATTTTTCAGAGCTCTCATTTT Mo17 GAAGAGCAGCAGTCTCTGCATGAAAGGGACTGAGGATGTCGCTCAGAAGCTATCACCTGATTTTTCAGAGCTCTCATTTT Consensus gaagagcagcagtctctgcatgaaagggactgaggatgtcgctca agctatcacctgatttttcagagctctcatttt 318 320 B73 AGGTTCTCTAAACTGCAGGTTTTCGTTGGCTAATATCGTTGGGTATCAAACTGAAACAGGTAGGGTGTAGCATCATGGTA Mo17 AGGTTGTCTAAACTGCAGGTTTTCGTTGGCTAATATCGTTGGGTATCAAACTGAAACAGGTAGGGTGTAGCATCATGGTA Consensus aggtt tctaaactgcaggttttcgttggctaatatcgttgggtatcaaactgaaacaggtagggtgtagcatcatggta 398 400 B73 GTTTGATTTCTGCTGTGGTGTTAGTTGGAGGGATAATGATTAGCGGCTAGTGGATTAAAGTTACCCATACCGTT Mo17 GTTTGATTTCTGCTGTGGTGTTAGTTGGAAGGATAATGATTAGCGGCTAGTGGATTAAAGTTACCCATACCGTT Consensus gtttgatttctgctgtggtgttagttgga ggataatgattagcggctagtggattaaagttacccataccgtt 472 474 Mei et al. B HuangC AGATCTCCTGCACCTTCGAACTCTGGAAAGATCATAGATTTTTGGGCAATAGTAAGTGGACATGGAACCATCCTCTCAGC 178 AGATCTCCTGCACCTTCGAACTCTGGAAAGATCATAGATTTTTGGGCAATAGTAAGTGGACATGGAACCATCCTCTCAGC Consensusagatctcctgcaccttcgaactctggaaagatcatagatttttgggcaatagtaagtggacatggaaccatcctctcagc 80 80 HuangC CTCAGCCTGCGATGGGTGTCGCCGCCGGTGGGTCACAAGTGTATCCTGCCTCTGCCTACCCGCCTGCAGCAACAGTAGCT 178 CTCAGCCTGCGATGGGTGTCGCCGCCGGTGGGTCACAAGTGTATCCTGCCTCTGCCTACCCGCCTGCAGCAACAGTAGCT Consensusctcagcctgcgatgggtgtcgccgccggtgggtcacaagtgtatcctgcctctgcctacccgcctgcagcaacagtagct 160 160 HuangC CCTCCTGCTGTTGCATCTGCTGGTTTACAGTCAGTGCAACCATTCCCAGCCAACCCTGCCCATATGAGTGCTCAGCACCA 178 CCTCCTGCTGTTGCATCTGCTGGTTTACAGTCAGTGCAACCATTCCCAGCCAACCCTGCCCATATGAGTGCTCAGCACCA Consensuscctcctgctgttgcatctgctggtttacagtcagtgcaaccattcccagccaaccctgcccatatgagtgctcagcacca 240 240 HuangC GATTGTCTACCAACAAGCTCAACAGTTCCACCAACAGCTCCAGCAGCAGCAACAGCAGCAGCTTCAGCAGTTCTGGGTCG 178 GATTGTCTACCAACAAGCTCAACAGTTCCACCAACAGCTCCAGCAGCAGCAACAGCAGCAGCTTCAGCAGTTCTGGGTCG Consensusgattgtctaccaacaagctcaacagttccaccaacagctccagcagcagcaacagcagcagcttcagcagttctgggtcg 320 320 HuangC AACGCATGACTGAAATCGAGGCAACAGCTGATTTCAGGAACCACAACTTGCCACTTGCGAGGATAAAGAAGATCATGAAG 178 AACGCATGACTGAAATCGAGGCAACAGCTGATTTCAGGAACCACAACTTGCCACTTGCGAGGATAAAGAAGATCATGAAG Consensusaacgcatgactgaaatcgaggcaacagctgatttcaggaaccacaacttgccacttgcgaggataaagaagatcatgaag 400 400 HuangC GCCGACGAAGATGTCCGCATGATCTCAGCCGAAGCTCCCGTGGTCTTCGCAAAAGCTTGCGAGATATTCATACTGGAGCT 178 GCCGACGAAGATGTCCGCATGATCTCAGCCGAAGCTCCCGTGGTCTTCGCAAAAGCTTGCGAGATATTCATACTAGAGCT Consensusgccgacgaagatgtccgcatgatctcagccgaagctcccgtggtcttcgcaaaagcttgcgagatattcatact gagct 480 480 HuangC GACGCTGAGGTCGTGGATGCACACCGAGGAGAACAAGCGCCGCACCTTGCAGAAGAACGACATTGCCGCAGCCATCACCA 178 GACGCTGAGGTCGTGGATGCACACCGAGGAGAACAAGCGCCGCACCTTGCAGAAGAACGACATTGCCGCAGCCATCACCA Consensusgacgctgaggtcgtggatgcacaccgaggagaacaagcgccgcaccttgcagaagaacgacattgccgcagccatcacca 560 560 HuangC GGACCGACATTTACGACTTCTTGGTCGACATTGTTCCCAGGGATGAGATGAAGGACGACGGAATCGGGCTTCCTAGGCCC 178 GGACCGACATTTACGACTTCTTGGTCGACATTGTTCCCAGGGATGAGATGAAGGACGACGGAATCGGGCTTCCTAGGCCC Consensusggaccgacatttacgacttcttggtcgacattgttcccagggatgagatgaaggacgacggaatcgggcttcctaggccc 640 640 HuangC GGGCTGCCACCCATGGGAGCCCCAGCTGACGCATATCCATACTACTACATGCCACAGCAGCAGGTGCCTGGTCCTGGGAT 178 GGGCTGCCACCCATGGGAGCCCCAGCTGACGCATATCCATACTACTACATGCCACAGCAGCAGGTGCCTGGTCCTGGGAT Consensusgggctgccacccatgggagccccagctgacgcatatccatactactacatgccacagcagcaggtgcctggtcctgggat 720 720 HuangC GGTTTATGGCGCCCAGCAAGGCCACCCCGTGACGTATCTGTGGCAGGATCCTCAGGAACAGCAGGAGCAAGCTCCCGAAG 178 GGTTTATGGCGCCCAGCAAGGCCACCCCGTGACGTATCTGTGGCAGGATCCTCAGGAACAGCAGGAGCAAGCTCCCGAAG Consensusggtttatggcgcccagcaaggccaccccgtgacgtatctgtggcaggatcctcaggaacagcaggagcaagctcccgaag 800 800 HuangC AGCAGCAGTCTCTGCATGAAAGGGACTGAGGATGTCGCACGTG 178 AGCAGCAGTCTCTGCATGAAAGGGACTGAGGATGTCGCACGTG Consensusagcagcagtctctgcatgaaagggactgaggatgtcgcacgtg 843 843 Figure S3 Sequence for CAPS analysis of NF-YC8. (A) Sequence for CAPS analysis of NF-YC8 in B73 and Mo17. This region contains a part of 3’ untranslated region. The SNP site was marked by red outline. (B) Sequence for CAPS analysis of NF-YC8 in Huang C and 178. The SNP site was marked by red outline. Mei et al. B73 CCCTTAGAGTTGATATAGCTCACTGACTTTTCAGCCTGTTCTTTTTACTGAAGAGCCATTGGCAATTTTGCCATTATCAT Mo17 CCTTTAGAGTTGATATAGCTCACTGACTTTTCAGCCTGTTCTTCTT.................................. Consensus cc ttagagttgatatagctcactgacttttcagcctgttctt tt 203 169 B73 TTGTGGGCTTCGCCAGTATGCCATCGGACCCACAAATCATAGACACAGACGGTCCCACCAGTCATAGACACGGGATGGCA Mo17 ................................................................................ Consensus 283 169 B73 TAATAACAAACAGCCAAAATTGAGAGTGGCAAAATAGCAAATTTCTCTTTTACTGAATGAGCAGGAGTGACGACAGGCTA Mo17 ................................................TTTACTGAATGAGCAGGAGTGACGACAGGCTA Consensus tttactgaatgagcaggagtgacgacaggcta 363 201 B73 CAGTTTCGGTCAGTGTCATCGGAGTCGGACGACAACCACGCGTACGTCCGCGACAAGGGCGATGACATGGTCGCAAGGTG Mo17 CAGTTTCGGTCAGTGTCATCGGAGTCGGACGACAACCACGCGTACGTCCGCGACAAGGGCGATGACATGGTCGCAAGGTG Consensus cagtttcggtcagtgtcatcggagtcggacgacaaccacgcgtacgtccgcgacaagggcgatgacatggtcgcaaggtg 443 281 B73 AAATTGAAGCCCGGCCTCATGGGCTGCGTCATGTGTGCATTTGCTGTGCTCGATTACTCGGAGGAGCATTTGCTTGCTTC Mo17 AAATTGAAGCCCGGCCTCATGGGCTGCGTCATGTGTGCATTTGCTGTGCTCGATTACTCGGAGGAGCATTTGCTTGCTTC Consensus aaattgaagcccggcctcatgggctgcgtcatgtgtgcatttgctgtgctcgattactcggaggagcatttgcttgcttc 523 361 B73 GTCCTTATCCTCCTTCCAAGACCAACCCTCGTATGCACGGTGACGTTAGCGCTAGGCTCTATTTGGAAGATGACTGATTC Mo17 GTCCTTATCCTCCTTCCAAGACCAACCCTCGTATGCACGGTGACGTTAGCGCTAGGCTCTATTTGGAAGATGACTGATTC Consensus gtccttatcctccttccaagaccaaccctcgtatgcacggtgacgttagcgctaggctctatttggaagatgactgattc 603 441 B73 TTGGTTTTTAAGGAACCGAGAAATCAGTTTTTAGAAATTGAATAAAAAAATTGACATATTTAGAGATCTTCCGGCTTTTA Mo17 TTGGTTTTTAAGGAACCGAGAAATCAGTTTTTAGAAATTGAATAAAAAA.TTGACATATTTAGAGATCTTCCGGCTTTTA Consensus ttggtttttaaggaaccgagaaatcagtttttagaaattgaataaaaaa ttgacatatttagagatcttccggctttta 683 520 B73 AAAACCAGTTTTCAAGAAACTAGATGTATCCAAACAAGTCTTAGACTATCTCCAGCAACGTCCTCTATATACATTATCTA Mo17 AAAACCAGTTTTCAAGAAACTAGATGTATCCAAACAAGTCTTAG.................................... Consensus aaaaccagttttcaagaaactagatgtatccaaacaagtcttag 763 564 B73 TATCCGTCTTTTACAGTCTCCTCTAAAAGATTTCATCTCCTATATCTACTTTCTCTTCAACAACGTCCTCTAAATCACGT Mo17 ................................................................................ Consensus 843 564 B73 CCTCTATATGTAAATACCTATATTAAAGACATTTTTAATTTTTTTAATTTTTGTACATACGTATTTGTCATACTCTCAAA Mo17 ................................................................................ Consensus 923 564 B73 TGTATTTTACATATTTTAGTTTTATTAAACCGGTTTAATAATAGATAGAGAACCGTTTAGAGAAACTCTACATATAGAGG Mo17 ................................................................................ Consensus 1003 564 B73 ATTCAGCAACGTTCTCTAAATTTAGAGGACCGTTTAGAGGACGTTGCTGGAGGGAGTAAAGGACGTCCTCGTCCTCTAAA Mo17 ................................................................................ Consensus 1083 564 B73 TTTAGGGTACAGGATCCTTTAGAGGGTCTTGTTGGAGCCAGCCTTAGTAGAGCTGCCCCTAGATTTTAAACTCACTTTTT Mo17 ...............................................TAGAGCTGCCCCTAGATTTTAAACTCACTTTTT Consensus tagagctgcccctagattttaaactcacttttt 1163 597 B73 AGGTTGCAAGGCAGTCATTTGCACAACCACGGTCCGTTTCCATGGCGACCCGCTCTCTGCCACCGACGCCCGGGCCCGCG Mo17 AGGTTGCAAGGCAGTCATTTGCACAACCACGGTCCGTTTCCATGGCGACCCGCTCTCTGCCACCGACGCCCGGGCCCGCG Consensus aggttgcaaggcagtcatttgcacaaccacggtccgtttccatggcgacccgctctctgccaccgacgcccgggcccgcg 1243 677 B73 GCCCAAACACCACCTGGACGGCTTGGGCCCCAGTACGGACCGGATTGAGGCCTGGCCTGGACCCGGAAGCACCAGGTGGT Mo17 GCCCAAACACCACCTGGACGGCTTGGGCCCCAGTACGGACCGGATTGAGGCCTGGCCTGGACCCGGAAGCACCAGGTGGT Consensus gcccaaacaccacctggacggcttgggccccagtacggaccggattgaggcctggcctggacccggaagcaccaggtggt 1323 757 B73 GCTGCCAATGGGACGCACGCTTTCGCTCCGAGGAGAATTTAAGAACCGAGTAGGCAGACAGGGAAGGCTAGGAGGGGACT Mo17 GCTGCCAATGGGACGCACGCTTTCGCTCCGAGGAGAATTTAAGAACCGAGTAGGCAGACAGGGAAGGCTAGGAGGGGACT Consensus gctgccaatgggacgcacgctttcgctccgaggagaatttaagaaccgagtaggcagacagggaaggctaggaggggact 1403 837 B73 AGGGGAGAGAGAGAG........GGGGAAGGCCAA Mo17 AGGGGAGAGAGAGAGAGAGAGAGGGGGAAGGCCAA Consensus aggggagagagagag ggggaaggccaa 1430 872 Figure S4 Sequence for methylation analysis of NF-YC8 5’ upstream region. The simple TE region was marked by red underline, repeat region was marked by blue underline. Mei et al. B73 GGTCACAAGTGTATCCTGCGTCTGCCTACCCGCCTGCAGCAACAGTAGCTCCTCCTGCTGTTGCATCTGCTGGTTTACAG Mo17 GGTCACAAGTGTATCCTGCCTCTGCCTACCCGCCTGCAGCAACAGTAGCTCCTCCTGCTGTTGCATCTGCTGGTTTACAG Consensus ggtcacaagtgtatcctgc tctgcctacccgcctgcagcaacagtagctcctcctgctgttgcatctgctggtttacag 80 80 B73 TCAGTGCAACCATTCCCAGCCAACCCTGCCCATATGAGTGCTCAGCACCAGATTGTCTACCAACAAGCTCAACAGTTCCA Mo17 TCAGTGCAACCATTCCCAGCCAACCCTGCCCATATGAGTGCTCAGCACCAGATTGTCTACCAACAAGCTCAACAGTTCCA Consensus tcagtgcaaccattcccagccaaccctgcccatatgagtgctcagcaccagattgtctaccaacaagctcaacagttcca 160 160 B73 CCAACAGCTCCAGCAGCAGCAACAGCAGCAGCTTCAGCAGTTCTGGGTCGAACGCATGACTGAAATCGAGGCAACAGCTG Mo17 CCAACAGCTCCAGCAGCAGCAACAGCAGCAGCTTCAGCAGTTCTGGGTCGAACGCATGACTGAAATCGAGGCAACAGCTG Consensus ccaacagctccagcagcagcaacagcagcagcttcagcagttctgggtcgaacgcatgactgaaatcgaggcaacagctg 240 240 B73 ATTTCAGGAACCACAACTTGCCACTTGCGAGGATAAAGAAGATCATGAAGGCCGACGAAGATGTCCGCATGATCTCAGCC Mo17 ATTTCAGGAACCACAACTTGCCACTTGCGAGGATAAAGAAGATCATGAAGGCCGACGAAGATGTCCGCATGATCTCAGCC Consensus atttcaggaaccacaacttgccacttgcgaggataaagaagatcatgaaggccgacgaagatgtccgcatgatctcagcc 320 320 B73 GAAGCTCCCGTGGTCTTCGCAAAAGCTTGCGAGATATTCATACTGGAGCTGACGCTGAGGTCGTGGATGCACACCGAGGA Mo17 GAAGCTCCCGTGGTCTTCGCAAAAGCTTGCGAGATATTCATACTGGAGCTGACGCTGAGGTCGTGGATGCACACCGAGGA Consensus gaagctcccgtggtcttcgcaaaagcttgcgagatattcatactggagctgacgctgaggtcgtggatgcacaccgagga 400 400 B73 GAACAAGCGCCGCACCTTGCAGAAGAACGACATTGCCGCAGCCATCACCAGGACCGACATTTACGACTTCTTGGTCGACA Mo17 GAACAAGCGCCGCACCTTGCAGAAGAACGACATTGCCGCAGCCATCACCAGGACCGACATTTACGACTTCTTGGTCGACA Consensus gaacaagcgccgcaccttgcagaagaacgacattgccgcagccatcaccaggaccgacatttacgacttcttggtcgaca 480 480 B73 TTGTTCCCAGGGATGAGATGAAGGACGACGGAATCGGGCTTCCTAGGCCCGGGCTGCCACCCATGGGAGCCCCAGCTGAC Mo17 TTGTTCCCAGGGATGAGATGAAGGACGACGGAATCGGGCTTCCTAGGCCCGGGCTGCCACCCATGGGAGCCCCAGCTGAC Consensus ttgttcccagggatgagatgaaggacgacggaatcgggcttcctaggcccgggctgccacccatgggagccccagctgac 560 560 B73 GCATATCCATACTACTACATGCCACAGCAGCAGGTGCCTGGTCCTGGGATGGTTTATGGCGCCCAGCAAGGCCACCCGGT Mo17 GCATATCCATACTACTACATGCCACAGCAGCAGGTGCCTGGTCCTGGGATGGTTTATGGCGCCCAGCAAGGCCACCCGGT Consensus gcatatccatactactacatgccacagcagcaggtgcctggtcctgggatggtttatggcgcccagcaaggccacccggt 640 640 B73 GACGTATCTGTGGCAGGATCCTCAGGAACAGCAGGAGCAAGCTCCTGAAGAG Mo17 GACGTATCTGTGGCAGGATCCTCAGGAACAGCAGGAGCAAGCTCCCGAAGAG Consensus gacgtatctgtggcaggatcctcaggaacagcaggagcaagctcc gaagag 692 692 Figure S5 Sequence for methylation analysis of NF-YC8 coding region B73 CTTGGTTTTGACTTTTGATGGGACCGCTTTAAGGAATTATGGGCCTGTTTGAGACAACTTTTTT.GGAAGAACTTTTCTG Mo17 GATGGCTTGG..TTTTGATGTGACTCCTTTAAGGAATTATGGGCCTGTTTGAGACAATTTTTTTTGAAAGAACTTTTCTG Consensustgg tt g ttttgatg gac ctttaaggaattatgggcctgtttgagacaa tttttt g aagaacttttctg 79 78 B73 AGAATATGAATTCTTGGATAATATAGCTGTCACGAGAATCTAGATATCACGAGGCTTCTATGCAGAGCAAGATAAAGGGG Mo17 AGAATATGAATTTTTGGATAATATAACTGTCACGAGAATCTGAATATCACGGGGCTTCTATGCAGAGCAAGTTAAAGGGG Consensus agaatatgaatt ttggataatata ctgtcacgagaatct atatcacg ggcttctatgcagagcaag taaagggg 159 158 B73 CATGCACAATTTAGGGTTTAAGAAGCCATAGATTCCTTGCGTCGCAACGACTCGATAAATTCTACATTTACGCGTTGAAT Mo17 CCTGCACAATTTAGGGTTTAAGAAGCGACAGATTCCTTGCGTCGCAACGACTCGACAAATTCTACATTCACGCGTTGAAT Consensus c tgcacaatttagggtttaagaagc a agattccttgcgtcgcaacgactcga aaattctacatt acgcgttgaat 239 238 B73 CAAGCGATTTTCACACATAAAAATGATTCTCACATAAACGGACTGAGAAAAGACGACTAGAATTCAACCAGAAGCCAAAA Mo17 CGAGTGATTTTCACACA..AAAATGATTCTCACATAAATGGACTGAGAAACGACGACTAGAATTTGACCAGAAGCCGAAA Consensus c ag gattttcacaca aaaatgattctcacataaa ggactgagaaa gacgactagaatt accagaagcc aaa 319 316 B73 CAAACAAGGACCATGTTATGTTATGCTACGTTT Mo17 CAAACAACGACCATGTTATGTTATGTTACACTT Consensus caaacaa gaccatgttatgttatg tac tt 352 349 Figure S6 Sequence (DMR) for methylation analysis of Mez1 (Haun et al. 2007) Mei et al. Figure S7 Bisulfite sequencing for the DMR of Mez1 5’ upstream region. (A, B) B73xMo17; (C, D) Mo17xB73. The DMR (350 bp) of the maternally expressed gene Mez1 5’ upstream was employed for verifying our bisulfite converted DNA templates. The specific-primers were used to amplify the DMR and the B73/Mo17 sequence polymorphisms allowed exact determination of the parental alleles for each clone. 9-15 bisulfite clones of each fragment were sequenced. Mei et al. Figure S8 Methylation analysis of the NF-YC8 5’ upstream region. (A-C) Methylation sites analysis of B73 alleles in endosperm tissues derived from B73xMo17 (maternal) and Mo17xB73 (paternal) reciprocal crosses at 18 DAP (TE region: -1244--1083; Repeat region: -686--273, differential methylated region was marked by red outline). (D-F) Methylation sites analysis of Mo17 alleles in endosperm tissues derived from Mo17xB73 (maternal) and B73xMo17 (paternal) reciprocal crosses at 18 DAP. Mei et al. Figure S9 Methylation analysis of the NF-YC8 coding region. (A, B) Methylation sites analysis of B73 alleles in endosperm tissues derived from B73xMo17 (maternal) and Mo17xB73 (paternal) reciprocal crosses at 18 DAP. (C, D) Methylation sites analysis of Mo17 alleles in endosperm tissues derived from Mo17xB73 (maternal) and B73xMo17 (paternal) reciprocal crosses at 18 DAP. Mei et al. Table S1 Methylated cytosine statistics of NF-YC8 in different alleles Table S1A Number of methylated cytosine in NF-YC8 5’ upstream of B73 allele Region 1 (-1287--1081) Number of clones CG (4 sites) CHG (8 sites) CHH (35 sites) B73xMo17_B73 allele 9 32/36 (88.89%) 57/72 (79.17%) 167/315 (53.02%) Mo17xB73_B73 allele 11 32/44 (72.73%) 83/88 (94.32%) 272/385 (70.65%) Region 2 (-1082--699) Number of clones CG (23 sites) CHG (14 sites) CHH (45 sites) B73xMo17_B73 allele 8 11/184 (5.98%) 15/112 (13.39%) 18/360 (5%) Mo17xB73_B73 allele 7 10/161 (6.21%) 14/98 (14.29%) 10/315 (3.17%) Region 3 (-698--519) Number of clones CG (4 sites) CHG (3 sites) CHH (36 sites) B73xMo17_B73 allele 12 43/48 (89.58%) 23/36 (63.89%) 64/432 (14.81%) Mo17xB73_B73 allele 14 196/196 (100%) 32/42 (76.19%) 183/504 (36.31%) Region 4 (-518--241) Number of clones CG (8 sites) CHG (8 sites) CHH (31 sites) B73xMo17_B73 allele 7 51/56 (91.07%) 25/56 (44.64%) 33/217 (15.21%) Mo17xB73_B73 allele 14 109/112 (97.32%) 41/112 (36.61%) 41/434 (9.44%) Region 5 (-240-+26) Number of clones CG (18 sites) CHG (19 sites) CHH (40 sites) B73xMo17_B73 allele 11 6/198 (3.03%) 12/209 (5.74%) 32/440 (7.27%) Mo17xB73_B73 allele 7 0/126 (0%) 3/133 (2.26%) 12/280 (4.29%) Mei et al. Table S1B Number of methylated cytosine in NF-YC8 5’ upstream of Mo17 allele Region 1 (-674--272) Number of clones CG (23 sites) CHG (15 sites) CHH (48 sites) B73xMo17_Mo17 allele 15 21/345 (6.09%) 40/225 (17.78%) 56/720 (7.78%) Mo17xB73_Mo17 allele 11 12/253 (4.74%) 25/165 (15.15%) 29/528 (5.49%) Region 2 (-271-+16) Number of clones CG (18 sites) CHG (19 sites) CHH (43 sites) B73xMo17_Mo17 allele 10 6/180 (3.33%) 8/190 (4.21%) 43/430 (10%) Mo17xB73_Mo17 allele 10 2/180 (1.11%) 9/190 (4.74%) 9/430 (2.09%) Table S1C Number of methylated cytosine in TE and repeat region of B73 allele TE region (-1244--1083) Number of clones CG (4 sites) CHG (5 sites) CHH (30 sites) B73xMo17_B73 allele 9 32/36 (88.89%) 40/45 (88.89%) 162/360 (45%) Mo17xB73_B73 allele 11 32/44 (72.73%) 55/55 (100%) 263/330 (79.70%) Repeat region (-686--273) Number of clones CG (12 sites) CHG (10 sites) CHH (58 sites) B73xMo17_B73 allele 7 76/84 (90.48%) 36/70 (51.43%) 67/406 (16.5%) Mo17xB73_B73 allele 14 165/168 (98.21%) 73/140 (52.14%) 196/812 (24.14%) Mei et al. Table S1D Number of methylated cytosine of NF-YC8 coding region Region 1 (+4080-+4421) Number of clones CG (12 sites) CHG (36 sites) CHH (58 sites) B73xMo17_B73 allele 7 76/84 (90.48%) 26/252 (10.32%) 12/406 (2.96%) Mo17xB73_B73 allele 8 89/96 (92.71%) 37/288 (12.85%) 13/472 (2.75%) Region 2 (+4422-+4771) Number of clones CG (20 sites) CHG (28 sites) CHH (51 sites) B73xMo17_B73 allele 6 66/120 (55%) 28/168 (16.67%) 12/306 (3.92%) Mo17xB73_B73 allele 9 130/180 (72.22%) 51/252 (20.24%) 20/459 (4.36%) Region 1 (+4080-+4421) Number of clones CG (11 sites) CHG (36 sites) CHH (60 sites) B73xMo17_Mo17 allele 5 40/55 (72.73%) 9/180 (5%) 4/300 (1.33%) Mo17xB73_Mo17 allele 6 60/66 (90.91%) 60/216 (27.78%) 19/360 (5.28%) Region 2 (+4422-+4771) Number of clones CG (21 sites) CHG (28 sites) CHH (51 sites) B73xMo17_Mo17 allele 7 87/ 147 (59.18%) 29/196 (14.80%) 12/357 (3.36%) Mo17xB73_Mo17 allele 7 93/147 (63.27%) 32/196 (16.33%) 14/357 (3.92%) CGG sites are counted as CG sites not as CHG. Asymmetric is defined by cytosine in the context CHH, where H=A, T, or C. Each fraction represents the number of cytosine methylated out of the total number of sites analyzed (Hermon et al. 2007). Mei et al. Table S2 Sequences of all PCR primers used Table S2A Primers for clone NF-YC8 clone NF-YC8F NF-YC8R NF-YC8 RACE 5'GSP-1 5'GSP-2 3'GSP-1 3'GSP-2 NF-YC8 CAPS analysis CAPS-F CAPS-R NF-YC8 RT-PCR RT-F RT-R NF-YC8 Promoter region Pr-F Pr-R NF-YC8 Intron region Fragment1F Fragment1R Fragment2F Fragment2R Mez1 Mez1F Mez1R NF-YC13 RT-PCR RT-F RT-R NF-YC14 RT-PCR RT-F RT-R TGCACCTTCGAACTCTGGAAA CGACATCCTCAGTCCCTTTCAT CCACGACCTCAGCGTCAGCTCCAGTAT CCAGCAGATGCAACAGCAGGAGGAGC GCTCCTCCTGCTGTTGCATCTGCTGG ATACTGGAGCTGACGCTGAGGTCGTGG ACCAGGACCGACATTTACGACTT AACGGTATGGGTAACTTTAATCCAC CGCATGACTGAAATCGAGGCAA AAGTCGTAAATGTCGGTCCTGGT CCCTTAGAGTTGATATAGCTCACTGACT CACTTCGAAGGTGCAGCAGAAGGT CATTCTCCAATCCGTGCCCTAGTC GTAAGGCCATTGACACATGTCCTC GAGGACATGTGTCAATGGCCTTAC GAGGATGGTTCCATGTCCACTTGC AGGAATCTTGATCCTACATTCTTC ATAACCTCGAGATAAAACAATCCC TAGGGCTGGTCTGCCACCCAT CACCCTACTTAGAATCGCCTGTTT TGGAGTAGACACGTCGGCTCAA TAGTCCAGAACTCCCTGAGCTG Mei et al. Table S2B Primers for methylation analysis Bisulfite sequencing for NF-YC8 Promoter region Bsr1-F1B Bsr1-R1B Bsr1-F2B(Second round) Bsr1-R2B(Second round) Bsr2-F1B Bsr2-R1B Bsr3-F1B Bsr3-R1B Bsr4-F1B Bsr4-R1B Bsr5-F1B Bsr5-R1B Bsr1-F1M Bsr1-R1M Bsr1-F2M(Second round) Bsr2-F1M Bsr2-R1M Coding region Bsr6-F1B Bsr6-F1M Bsr6-R1 Bsr7-F1 Bsr7-R1B Bsr7-R1M Bisulfite sequencing for Mez1 BSr-F1B BSr-F2B(Second round) BSr-R1B BSr-F1M BSr-F2M(Second round) BSr-R1M TTAGAGTTGATATAGTTTAYTGAYTTTTYAGTYTG CTRTCRTCACTCCTRCTCATTCARTA GTTGATATAGTTTAYTGAYTTTTYAGTYTGTT ACCRAAACTRTAACCTRTCRTCAC TAYTGAATGAGYAGGAGTGAYGAYAG AACRTTRCTRAAAATAATCTAARACTTRTTTRAAT ATTYAAAYAAGTYTTAGATTATTTTYAGYAAYGTT AAATACATTTRAAAATATAACAAATACRTATATAC GTATATAYGTATTTGTTATATTTTYAAATGTATTT TAATTRTACAAATAACTRCCTTRCAACCT AGGTTGYAAGGYAGTTATTTGTAYAATTA TTRACCTTCCCCCTCTCTCTCTCC ATGTATTYAAAYAAGTRTTAGTAGAGYTGTT TCTAAAAACARCTCTACTAARACTTRTTTAAAT TTAYTGAATGAGYAGGAGTGAYGA ATTTAAAYAAGTYTTAGTAGAGYTGTTTTTAGA CCTCTCTCTCTCTCTCTCTCC GGTTATAAGTGTATTYTGYGTYTGTTTAT GGTTATAAGTGTATTYTGTTTYTGTTTAT TATCTCRCAAACTTTTACRAAAACCA AAGTTTGYGAGATATTTATAYTGGAGTT CTCTTCARAAACTTACTCCTRCTRTT CTCTTCRRAAACTTACTCCTRCTRTT TTATTTTTTYTGTGTTYAAAGGGGGTTA TTGGTTTTGATTTTTGATGGGA AAACRTAACATAACATAACATAATCCTT TTGTTTTTTGYAATTYAAAGGGGGTTAA GATGGTTTGGTTTTGATGTGA AAATATAACATAACATAACATAATCRTT Mei et al. Table S3 IDs for Arabidopsis thaliana NF-YC subfamily (Siefers et al. 2009) NF-YC1 NF-YC2 NF-YC3 NF-YC4 NF-YC5 NF-YC6 NF-YC7 NF-YC8 NF-YC9 NF-YC10 NF-YC11 NF-YC12 NF-YC13 The protein sequences of AT3G48590 AT1G56170 AT1G54830 AT5G63470 AT5G50490 AT5G50480 AT5G50470 AT5G27910 AT1G08970 AT1G07980 AT3G12480 AT5G38140 AT5G43250 Arabidopsis thaliana NF-YC subfamily were downloaded from http://ensembl.gramene.org/Arabidopsis_thaliana/Info/Index. Reference Haun WJ, Laoueille-Duprat S, O'Connell M J, Spillane C, Grossniklaus U, Phillips AR, Kaeppler SM, Springer NM (2007) Genomic imprinting, methylation and molecular evolution of maize Enhancer of zeste (Mez) homologs. The Plant journal : for cell and molecular biology 49: 325-337 Hermon P, Srilunchang KO, Zou J, Dresselhaus T, Danilevskaya ON (2007) Activation of the imprinted Polycomb Group Fie1 gene in maize endosperm requires demethylation of the maternal allele. Plant molecular biology 64: 387-395 Liang M, Yin X, Lin Z, Zheng Q, Liu G, Zhao G (2014) Identification and characterization of NF-Y transcription factor families in Canola (Brassica napus L.). Planta 239: 107-126 Lin YX, Jiang HY, Chu ZX, Tang XL, Zhu SW, Cheng BJ (2011) Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC genomics 12: 76 Petroni K, Kumimoto RW, Gnesutta N, Calvenzani V, Fornari M, Tonelli C, Holt BF, III, Mantovani R (2012) The Promiscuous Life of Plant NUCLEAR FACTOR Y Transcription Factors. The Plant cell 24: 4777-4792 Siefers N, Dang KK, Kumimoto RW, Bynum WE, Tayrose G, Holt BF, III (2009) Tissue-Specific Expression Patterns of Arabidopsis NF-Y Transcription Factors Suggest Potential for Extensive Combinatorial Complexity. Plant physiology 149: 625-641 Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution 28: 2731-2739 Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic acids research 25: 4876-4882