tpj13125-sup-0015-Legend
advertisement

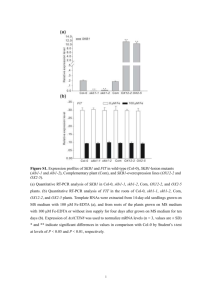
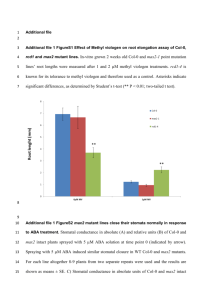
Supporting Information Legends Figure S1. Sequence alignment showing that Arabidopsis SBB1 is similar to NUP85 homologs from rice, human and yeast. Figure S2. RT-PCR analyses show the genetic backgrounds of two T-DNA insertion alleles of sbb1. Figure S3. The genetic background of pSBB1::SBB1g transgenic plants and the corresponding mutants were verified by RT-PCR analyses. Figure S4. bak1-3 bkk1-1 sbb1 is less sensitive to SA than bak1-3 bkk1-1. Seedlings were grown on ½ MS media supplemented with 0, 10 and 50 μM SA respectively and grown for two weeks. Phenotypes of representative seedlings for different treatments are shown. Scale bars represent 1 mm. Figure S5. Loss-of-function mutations of NUP133, NUP107, NUP43 and SEC13B cannot fully suppress the cell-death phenotype of bak1-3 bkk1-1. (a-d) Cell-death phenotype of bak1-3 bkk1-1 is not fully suppressed by knocking-out (a) NUP133, (b) NUP107, (c) NUP43 or (d) SEC13B (three-week-old). (e-t) Trypan blue staining of the cotyledons show cell-death symptoms of bak1-3 bkk1-1 are almost not rescued by (e-h) nup133, (i-l) nup107, (m-p) nup43 and (q-t) sec13b. (e) Col-0; (f) bak1-3 bkk1-1; (g) nup133-3; (h) bak1-3 bkk1-1 nup133-3; (i) Col-0; (j) bak1-3 bkk1-1; (k) nup107-1; (l) bak1-3 bkk1-1 nup107-1; (m) Col-0; (n) bak1-3 bkk1-1; (o) nup43-1; (p) bak1-3 bkk1-1 nup43-1. (q) Col-0; (r) bak1-3 bkk1-1; (s) sec13b-1; (t) bak1-3 bkk1-1 sec13b-1. (u-x) Relative expression of PR1, PR2 and PR5, and SA levels in various genotypes. Scale bars represent 1 cm (a-d) and 200 μm (e-t). Figure S6. RT-PCR analyses show the genetic backgrounds of various mutants. Figure S7. DRH1 localizes in the nucleus. (a) Transient expression of 35S-DRH1-YFP in an Arabidopsis protoplast, (b) bright field, (c) image merge of 35S-DRH1-YFP and bright field. Scale bars represent 10 μm. Figure S8. DRH1 physically interacts with NUP107 by yeast two-hybrid. Figure S9. mRNA export is affected in nup133, nup107, nup43 mutants but not in sec13b. (a-h) Whole mount in situ hybridization of mRNA shows that poly(A)+ RNAs are accumulated more or less in the nuclei of nup133-3, nup107-1, nup43-1 single or triple mutant, but uniformly distributed in the nuclei and cytoplasm of sec13b-1 single or triple mutant. (a) nup133-3, (b) bak1-3 bkk1-1 nup133-3, (c) nup107-1, (d) bak1-3 bkk1-1 nup107-1, (e) nup43-1, (f) bak1-3 bkk1-1 nup43-1, (g) sec13b-1, (h) bak1-3 bkk1-1 sec13b-1. Scale bars represent 20 μm (a-h). Figure S10. Nuclear accumulation of the mRNAs of several genes that regulate SA biosynthesis are not obviously affected in the sbb1-1 and drh1-1 mutants comparing Col-0. qRT-PCR analyses of EDS1, EDS5, PAD4 and SID2 using RNA from cytoplasmic and nuclear fraction or total RNA of 7-day-old Col-0, sbb1-1, drh1-1 seedlings. ACT2 was used as internal control for the qRT-PCR analyses. Figure S11. Cell death of bir1 and bon1 cannot be suppressed by SBB1 mutation. (a) Phenotypes of 16-day-old of sbb1 and bir1 related seedlings grown in the soil. (b-e) Trypan blue staining showing that cell death of bir1 was not able to be rescued by sbb1. (b) Col-0, (c) sbb1-1, (d) bir1-1, (e) bir1-1 sbb1-1. (f) Phenotypes of 3-week-old of sbb1 and bon1 related seedlings grown in the soil. (g-j) Trypan blue staining showing that SBB1 mutation cannot suppress the cell-death phenotype of bon1. (g) Col-0, (h) sbb1-1, (i) bon1-1, (j) bon1-1 sbb1-1. Scale bars represent 1 cm (a and f) and 1mm (b-e, g-j). Method S1. DRH1 cloning and transient expression in protoplast of Arabidopsis leaves Full length CDS of DRH1 was amplified by regular RT-PCR and recombined into binary vector pUC19-35S-GWR-YFP using a Gateway® cloning approach (Invitrogen). Primers used for amplifying CDS of DRH1 are 5’-AAAAAGCAGGCTTCATGGCTGCTACCGCTGCTGCTTC-3’, 5’-AGAAAGCTGGGTCTCTGTGTTTCATCATCATCGTCTCG-3’. pUC19-35S-DRH1-YFP was transformed into Arabidopsis leaf protoplasts according to previous studies (Yoo et al. 2007). YFP fluorescence signals were observed under a confocal microscope. Method S2. Yeast two-hybrid assay Yeast two-hybrid analyses was performed using the ProQuest™ Two-Hybrid System with Gateway® Technology according to the manual. Supporting reference Yoo, S.-D., Cho, Y.-H. and Sheen, J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protocols, 2, 1565-1572. Appendix S1. A proteomic approach was used to discover new components involving in SBB1 controlled cell-death signaling pathways. A number of SBB1-interacting proteins including DEAD box RNA helicases were identified by immunoprecipitation and LC-MS/MS analyses.