tpj12995-sup-0007-FigLegend
advertisement

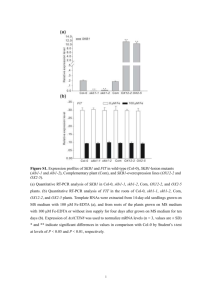
Zamioudis et al. – Page 1/3 FULL LEGENDS SUPPORTING INFORMATION Figure S1. Quantitative ferric-chelate reductase activity in roots of Arabidopsis Col-0 seedlings that were grown under conditions of sufficient iron (50 µM Fe(III)EDTA) and were either mocktreated (Control) or treated with the ISR-inducing rhizobacterial Pseudomonas strains WCS417 or WCS358 or the ISR-noninducing Pseudomonas strain WCS374. Iron deprived plants (-Fe, 300 μΜ Ferrozine) were used as positive control. Seedlings were treated when 12 days old and samples were collected 2 days later. Data represent the mean of three replicates (±SD). Different letters indicate statistically significant differences (Tukey’s HSD test; p<0.05). Figure S2. Representative confocal images of pMYB72:GFP-GUS roots of 2-week-old seedlings that were grown under conditions of sufficient iron (50 µM Fe(III)EDTA) and were either mock-treated (Control) or treated with the ISR-inducing rhizobacterial Pseudomonas strains WCS417 or WCS358 or the ISR-noninducing Pseudomonas strain WCS374. Images were taken 2 days after treatment. LMZ: late maturation zone. Scale bars: 50 µm. Figure S3. Bacterial determinants involved in the induction of the iron deficiency response in Arabidopsis roots. (a) Quantitative RT-PCR analysis of MYB72 transcripts in roots of 12-day-old Col-0 seedlings that were grown under conditions of sufficient iron (50 µM Fe(III)EDTA), 2 days after treatment with MgSO4 (Control), wild-type P. simiae WCS417 bacteria, or the siderophoreimpaired mutants WCS417-S680 or WCS417-M634. (b) Quantitative RT-PCR analysis of CYP71A12 (MAMP-responsive positive control), MYB72 and FRO2 transcript levels in the roots of 2-week-old Col-0 seedling that were either mock-treated (Control) or treated with 250 nM flg22 or 0.001% chitosan. Samples were collected for gene expression analysis at the indicated time points after treatment. Data represent the mean of three replicates (±SD). Asterisks Zamioudis et al. – Page 2/3 indicate statistically significant differences compared with mock-treated roots (Student’s t test; p<0.05). Figure S4. Bacterial VOCs activate the Strategy I iron deficiency response independent of the iron status in the root vicinity. Quantitative RT-PCR analysis of (a) MYB72 and (b) FRO2 transcripts in the roots of mock-treated (Control) and P. simiae WCS417-VOCs-treated Arabidopsis Col-0 seedlings. Twelve-day-old seedlings growing for 5 days in modified Hoagland medium supplemented with the indicated concentrations of Fe(III)EDTA were treated with WCS417-VOCs and samples were collected 24 h later. Numbers above bars indicate the absolute fold-induction level in gene expression under the respective concentrations of iron. Table S1. Up- and down-regulated genes in Arabidopsis Col-0 roots in response to colonization by P. simiae WCS417 bacteria (p<0.05 and >1.5 fold-change). Two-week-old Arabidopsis Col-0 seedlings growing in modified Hoagland agar medium were inoculated with WCS417 by applying 10-μl bacterial suspension (107 cfu mL-1) to the primary root of each seedling. Forty eight h later, three biological replicates of mock- and WCS417-treated roots were harvested for microarray analysis. Table S2. Selected members of the Arabidopsis natural root microbiome activate MYB72 expression. Twelve-day-old pMYB72:GFP-GUS seedlings were exposed for 2 days to VOCs of the indicated bacterial strains in a split-plate assay and the intensity of GUS staining in the roots was evaluated compared to mock-treated seedlings in the order of increasing activity: + < ++ < +++. Values in parentheses indicate intermediate intensity levels. P. simiae strain WCS417 was used as a reference. ID: strain number; OTU: operational taxonomic unit number as published previously (Bulgarelli et al. 2012). Zamioudis et al. – Page 3/3 Table S3. Arabidopsis Genome Initiative numbers and primers used in this study for cloning and studying expression of Arabidopsis genes.