tpj12996-sup-0005-FigLegends
advertisement

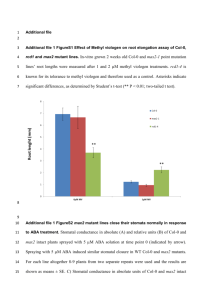
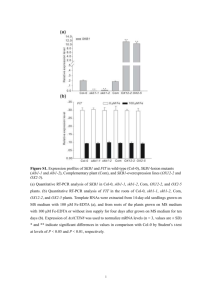
Legends for all the Supporting Material Supporting Figures Figure S1. Phenotypes of A. thaliana Col-0 and bzip19-1 mutant grown on MGRL- and MS-based Zn-deficient medium solidified with purified agar or general agar. (a) MGRL, Purified agar(corresponded to Zn0 medium used in this study), (b) MGRL, General agar, (c) MS, Purified agar, (d) MS, General agar. Horizontal bar, 20 mm. Figure S2. Root length of A. thaliana Col-0 and mutants of bZIP19 and bZIP23 grown on Zn10, Basal, and Zn0 media (means ± SEM; n=40). These phenotypes are shown in Figure 1c. Figure S3. Relative transcript levels of ZIP7, 8, 10, and 11 in the root of A. thaliana Col-0 (Col), bzip19-1 (b19), and bzip23-1 (b23) mutants grown on Zn10 (black bars), Basal (grey bars), and Zn0 (white bars) media (means ± SD). Expression of each gene in Col-0/Basal was set to 1 as the frame of reference. Figure S4. Comparison of the relative transcript levels of ZIP family members in the roots of A. thaliana Col-0 grown on Basal medium. Expression of ZIP1 was set to 1 as 1 the frame of reference. Figure S5. ZDRE motif in the promoter regions of defensin-like genes, ZIP9 and ZIP3. Boxes show TCGA core sequence. Crosses above the sequence show mismatches from the ZDRE consensus sequence (RTGTCGACAY). Negative integers above the sequence show nucleotide distance from the ATG start codon. Supporting Table Table S1. List of all proteins identified by iTRAQ analysis. Only proteins identified and quantified more than five times in eight independent experiments in any condition are listed. Peptides from microsomal proteins of Col-0 roots on Basal medium, Col-0 roots on Zn0 medium, bzip19-1 mutant roots on Basal medium and bzip19-1 mutant roots on Zn0 medium were labeled with iTRAQ-114 (Col-0 Basal), 115 (Col-0 Zn0), 116 (bzip19-1 Basal) and 117 (bzip19-1 Zn0) reagent, respectively. The values were calculated as the ratio of label in 115, 116 and 117 to 114 or 117 to 116. Data shown represent means ± SD. Proteins were ordered based on AGI Code numbers. “TM Domain” indicates the number of TM regions predicted by the 2 TMHMM Server version 2.0. "Count" indicates number of times identified and quantified peptides in LC-MS analysis. Table S2. Measured concentrations (mM) of micro- and macro- elements in the growth media Data are shown as concentration per volume (mean ± SD; n=3). Supporting Method Method S1. Phenotype of bzip19-1 mutant on different type of Zn-deficient media Method S2. iTRAQ analysis 3