Questions
advertisement

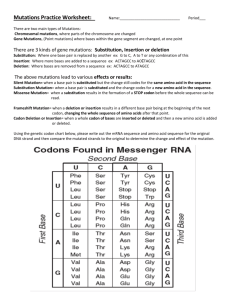
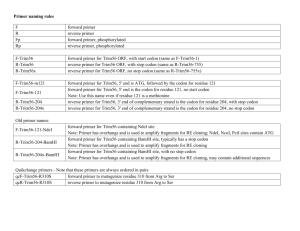
Questions 1. Give examples of cases where site-directed mutagenesis can be useful. The applicatons of site-directed mutagenesis can generall be divided into two categories: 1. Tools for mapping the functions and properties of a DNA sequence, an RNA sequence or a protein. 2. Rational design of proteins. By introducing a specific mutation in DNA and investigating the effect of this mutation on the DNA sequence or its product, functions and properties may be deduced. Site-directed mutations may also be introduced in order to tailor the properties of properties based on theoretical analyses. 2. The DNA-sequence encoding the NNHSYW-motif in algL can also also be mutated to sequence corresponding to NNASYW. Suggest a QuikChange primerpair that changes the NNHSYW to NNASYW. The mutagenic primers should 1) encode the NNASYW-motif in the right reading frame and 2) contain a new restriction site for identification of the mutaion (find an alternative to BpmI!). Be careful to only introduce “silent mutations”, except for the intended H→A-mutation. The length of the primer should be ~45 nucleotides and the mutations should be as close to each other and as centered in the primer as possible (…). The wild-type sequence with amino acid sequence for the correct reading frame: acctgccgct ggaaaaaacc aacaaccact cctactgggc cgcctggtcg l p l e k t n n h s y w a a w s Histidine (h) in the wild-type sequence should be changed to alanine (a). Alanine is encoded by all codons starting with gc, i.e. gct, gcc, gca and gcg. In this case, the simplest approach is to change the first two nucleotides in the histidine codon from ca to cg, thereby changing the codon from cag to gcc. Furthermore, the primers should a new restriction site introduced throug a silent mutation, i.e. without changing the amino acid sequence of the protein. There are several ways to achieve this. One example is: acctgccgct Cgaaaaaacc aacaaccact cctactgggc cgcctggtcg Here, a mutation from g to c has been introduced in nucleotide number 11 from the left (signified by an uppercase letter). The new codon, ctc, encodes leucine, just like the original codon, ctg. The underlined sequence is the restriction site of the enzyme AccBSI [1]. Another exampe is: acctgccAct ggaaaaaacc aacaaccact cctactgggc cgcctggtcg This mutation changes nucleotide number 8 from the left from g to a. The codon is thereby changed from ccg to cca; both of these encode proline. The underlined sequence is the restriction site of Bse1I [2]. Here, it is chosen to utilise the first example to introduce a restriction site. The desired mutant sequence then becomes: acctgccgct Cgaaaaaacc aacaacGCct cctactgggc cgcctggtcg l p l e k t n n A s y w a a w s Mutations that are introduced in the DNA sequence as wall as the resulting mutation in the protein product are signified by uppercase letters. The restriction site of AccBSI is underlined. The following mutagenesis primers can be used to achieve this: Forward primer: 5’-cctgccgctcgaaaaaaccaacaacgcctcctactgggccgcctg-3’ Reverse primer: 5’-caggcggcccagtaggaggcgttgttggttttttcgagcggcagg-3’ The forward primer contains the desired mutations and is otherwise identical to the original wild-type sequence. The reverse primer is complementary to the forward primer and thus contains mutations that are complementary to those introduced in the forward primer. Both primers have a length of 45 bp. References 1. http://rebase.neb.com/rebase/enz/AccBSI.html 2. http://rebase.neb.com/rebase/enz/Bse1I.html