Protocol for Making a New Construct By PCR
advertisement

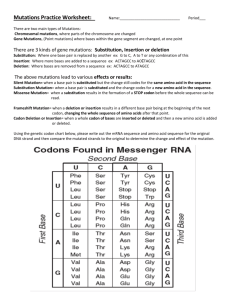
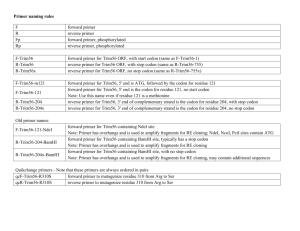
Protocol for Making a New Construct By PCR 1) What is the sequence you are trying to amplify/clone? - Don’t trust database sequences to be correct; they often contain extra amino acids that were useful to a particular construct, but that you might not necessarily want (look for the reading frame, not just the insert) - Compare the sequence in the database to SwissProt sequence information: do they match? - If only amplifying a domain, the SwissProt version of that domain may be different than the one you want: be sure of what is needed for functionality 2) What primers/restriction sites should be used to amplify the sequence? - Start by finding two appropriate restriction sites within the vector that DO NOT have complementary sticky ends - Check the NEB catalog and add bases to the 5’ (exposed) ends as necessary to increase the efficiency of the respective enzymes. It is not necessary to add bases to both sides of the site, only the side that is NOT surrounded by the rest of the amplified fragment. - In general, you want the primer to have a Tm of about 60C, and a GC content of about 50.0%, but in practice this is rarely possible, especially when the sequence to be amplified cannot be altered. - It is always good to pin down the 3’ end of any primer with a C or G, but not absolutely necessary, especially when you are just amplifying of a plasmid. 3) Have I done this right? - Check the reading frame by constructing a predicted sequence: find your enzyme sites within the vector sequence, delete the base pairs in the middle, and then paste in your sequence (only the sequence that would be amplified by your primers). - Translate the entire open reading frame and search for the first few amino acids of your fluorescent protein or transgene (whichever comes last): if things are in frame, you will find these residues. - If the fusion is not in frame, add bases to the end of the appropriate primer until it is (either one or two bases). It is best to use bases that result in neutral amino acids in the linker (glycine and alanine are best). 4) Random but important points - Do you have a Kozak sequence (ACC) just before the start codon? - Do you have a start codon (ATG)? - Do you have a stop codon where you shouldn’t (ie. at the end of a transgene where it is fused to the N terminus of the fluorescent tag). - Do you have a stop codon where you should (ie. at the end of the transgene where it is fused to the C terminus of the fluorescent tag).